Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model
- PMID: 17442505
- PMCID: PMC1950257
- DOI: 10.1016/j.gene.2007.02.026
Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model
Abstract
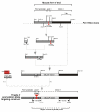
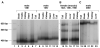
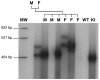
Carriers of FMR1 alleles with 55-200 repeats in the 5' UTR are at risk for Fragile X associated tremor and ataxia syndrome. The cause of the neuropathology is unknown but is thought to be RNA-mediated. Maternally transmitted premutation alleles are also at risk of expansion of the repeat tract into the "full mutation" range (>200 repeats). The mechanism responsible for expansion is unknown. Full mutation alleles produce reduced amounts of the FMR1 gene product, FMRP, which leads to Fragile X mental retardation syndrome. We have developed a murine model for Fragile X premutation carriers that recapitulates key features seen in humans including a direct relationship between repeat number and Fmr1 mRNA levels, an inverse relationship with FMRP levels and Purkinje cell dropout that have not been seen in a previously described knock-in mouse model. In addition, these mice also show a differential deficit of FMRP in different parts of the brain that might account for symptoms of the full mutation that are seen in premutation carriers. As in humans, repeat instability is high with expansions predominating and, for the first time in a mouse model, large expansions into the full mutation range are seen that occur within a single generation. Thus, contrary to what was previously thought, mice may be good models not only for the symptoms seen in human carriers of FMR1 premutation alleles but also for understanding the mechanism responsible for repeat expansion, a phenomenon that is responsible for a number of neurological and neurodevelopmental disorders.
Figures





References
-
- Aziz M, Stathopulu E, Callias M, Taylor C, Turk J, Oostra B, Willemsen R, Patton M. Clinical features of boys with fragile X premutations and intermediate alleles. Am J Med Genet B Neuropsychiatr Genet. 2003;121:119–27. - PubMed
-
- Bontekoe CJ, Bakker CE, Nieuwenhuizen IM, van der Linde H, Lans H, de Lange D, Hirst MC, Oostra BA. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum Mol Genet. 2001;10:1693–9. - PubMed
-
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–87. - PubMed
-
- Dutch-Belgian Fragile X Consortium Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

