Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins
- PMID: 17446074
- PMCID: PMC1885954
- DOI: 10.1016/j.cub.2007.03.064
Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins
Abstract
Background: The nucleolus is a subnuclear organelle in which rRNAs are transcribed, processed, and assembled with ribosomal proteins into ribosome subunits. Mass spectrometry combined with pulsed incorporation of stable isotopes of arginine and lysine was used to perform a quantitative and unbiased global analysis of the rates at which newly synthesized, endogenous proteins appear within mammalian nucleoli.
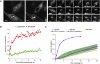
Results: Newly synthesized ribosomal proteins accumulated in nucleoli more quickly than other nucleolar components. Studies involving time-lapse fluorescence microscopy of stable HeLa cell lines expressing fluorescent-protein-tagged nucleolar factors also showed that ribosomal proteins accumulate more quickly than other components. Photobleaching and mass-spectrometry experiments suggest that only a subset of newly synthesized ribosomal proteins are assembled into ribosomes and exported to the cytoplasm. Inhibition of the proteasome caused an accumulation of ribosomal proteins in the nucleus but not in the cytoplasm. Inhibition of rRNA transcription prior to proteasomal inhibition further increased the accumulation of ribosomal proteins in the nucleoplasm.
Conclusions: Ribosomal proteins are expressed at high levels beyond that required for the typical rate of ribosome-subunit production and accumulate in the nucleolus more quickly than all other nucleolar components. This is balanced by continual degradation of unassembled ribosomal proteins in the nucleoplasm, thereby providing a mechanism for mammalian cells to ensure that ribosomal protein levels are never rate limiting for the efficient assembly of ribosome subunits. The dual time-lapse strategy used in this study, combining proteomics and imaging, provides a powerful approach for the quantitative analysis of the flux of newly synthesized proteins through a cell organelle.
Figures






Comment in
-
Building ribosomes: even more expensive than expected?Curr Biol. 2007 Jun 5;17(11):R415-7. doi: 10.1016/j.cub.2007.04.011. Curr Biol. 2007. PMID: 17550767
References
-
- Mayer C., Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. - PubMed
-
- Lewis J.D., Tollervey D. Like attracts like: Getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. - PubMed
-
- Tate W.P., Poole E.S. The ribosome: Lifting the veil from a fascinating organelle. Bioessays. 2004;26:582–588. - PubMed
-
- Granneman S., Baserga S.J. Ribosome biogenesis: Of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. - PubMed
-
- Mann M. Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 2006;7:952–958. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

