Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation
- PMID: 17446867
- PMCID: PMC1864975
- DOI: 10.1038/sj.emboj.7601677
Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation
Abstract
On the basis of kinetic data on ribosome protein synthesis, the mechanical energy for translocation of the mRNA-tRNA complex is thought to be provided by GTP hydrolysis of an elongation factor (eEF2 in eukaryotes, EF-G in bacteria). We have obtained cryo-EM reconstructions of eukaryotic ribosomes complexed with ADP-ribosylated eEF2 (ADPR-eEF2), before and after GTP hydrolysis, providing a structural basis for analyzing the GTPase-coupled mechanism of translocation. Using the ADP-ribosyl group as a distinct marker, we observe conformational changes of ADPR-eEF2 that are due strictly to GTP hydrolysis. These movements are likely representative of native eEF2 motions in a physiological context and are sufficient to uncouple the mRNA-tRNA complex from two universally conserved bases in the ribosomal decoding center (A1492 and A1493 in Escherichia coli) during translocation. Interpretation of these data provides a detailed two-step model of translocation that begins with the eEF2/EF-G binding-induced ratcheting motion of the small ribosomal subunit. GTP hydrolysis then uncouples the mRNA-tRNA complex from the decoding center so translocation of the mRNA-tRNA moiety may be completed by a head rotation of the small subunit.
Figures
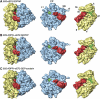


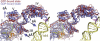


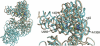
References
-
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol 6: 643–647 - PubMed
-
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J (2005) The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712 - PubMed
-
- Armstrong S, Yates SP, Merrill AR (2002) Insight into the catalytic mechanism of Pseudomonas aeruginosa exotoxin A. Studies of toxin interaction with eukaryotic elongation factor-2. J Biol Chem 277: 46669–46675 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

