In vivo gene delivery and expression by bacteriophage lambda vectors
- PMID: 17448169
- PMCID: PMC2063594
- DOI: 10.1111/j.1365-2672.2006.03182.x
In vivo gene delivery and expression by bacteriophage lambda vectors
Abstract
Aims: Bacteriophage vectors have potential as gene transfer and vaccine delivery vectors because of their low cost, safety and physical stability. However, little is known concerning phage-mediated gene transfer in mammalian hosts. We therefore performed experiments to examine phage-mediated gene transfer in vivo.
Methods and results: Mice were inoculated with recombinant lambda phage containing a mammalian expression cassette encoding firefly luciferase (luc). Efficient, dose-dependent in vivo luc expression was detected, which peaked within 24 h of delivery and declined to undetectable levels within a week. Display of an integrin-binding peptide increased cellular internalization of phage in vitro and enhanced phage-mediated gene transfer in vivo. Finally, in vivo depletion of phagocytic cells using clodronate liposomes had only a minor effect on the efficiency of phage-mediated gene transfer.
Conclusions: Unmodified lambda phage particles are capable of transducing mammalian cells in vivo, and may be taken up -- at least in part -- by nonphagocytic mechanisms. Surface modifications that enhance phage uptake result in more efficient in vivo gene transfer.
Significance and impact of the study: These experiments shed light on the mechanisms involved in phage-mediated gene transfer in vivo, and suggest new approaches that may enhance the efficiency of this process.
Figures




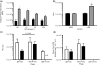
 ) phage (containing a modified gpD protein bearing the 3JCLI4 integrin-binding peptide) at an MOI of 105, in the presence or absence of increasing concentrations of soluble 3JCLI4 protein. Two hours later, cells were washed and lysates were prepared in order to quantify internalized phage, which were then titrated on LE392 Escherichia coli cells. Data shown represent mean phage titres ± SDs (calculated from three independently analysed wells of a cell culture dish, each of which was titrated in triplicate). There was a statistically significant increase in phage internalization between the 3JCLI4 (luc) phage particles and the gpD (luc) phage particles (*P < 0·05, two-way
) phage (containing a modified gpD protein bearing the 3JCLI4 integrin-binding peptide) at an MOI of 105, in the presence or absence of increasing concentrations of soluble 3JCLI4 protein. Two hours later, cells were washed and lysates were prepared in order to quantify internalized phage, which were then titrated on LE392 Escherichia coli cells. Data shown represent mean phage titres ± SDs (calculated from three independently analysed wells of a cell culture dish, each of which was titrated in triplicate). There was a statistically significant increase in phage internalization between the 3JCLI4 (luc) phage particles and the gpD (luc) phage particles (*P < 0·05, two-way  ) phage was added to K562-αvβ3 or wild-type K562 cells in 96-well plates, at an MOI of 105. The plates were subjected to centrifugation at 900 g for 15 min, in order to enhance the efficiency of phage binding to the target cells (O'Doherty et al. 2000; Scanlan et al. 2005; Harui et al. 2006a). After this, the cultures were returned to a 37°C incubator for 1 h and 45 min, and the cells were then washed to remove unbound phage. The cultures were again returned to a 37°C incubator, and 48 h later, cell lysates were prepared. After normalization of the protein content of the cell lysates, luc expression was measured. Data shown represent mean luc expression values ± SDs (calculated from three independent experiments, each of which analysed triplicate wells of a cell culture dish). There was a statistically significant increase in luc expression between the 3JCLI4 (luc) phage particles and the gpD (luc) phage particles, when tested in K562-αvβ3 cells but not when tested in wild-type K562 cells (*P < 0·05, two-way
) phage was added to K562-αvβ3 or wild-type K562 cells in 96-well plates, at an MOI of 105. The plates were subjected to centrifugation at 900 g for 15 min, in order to enhance the efficiency of phage binding to the target cells (O'Doherty et al. 2000; Scanlan et al. 2005; Harui et al. 2006a). After this, the cultures were returned to a 37°C incubator for 1 h and 45 min, and the cells were then washed to remove unbound phage. The cultures were again returned to a 37°C incubator, and 48 h later, cell lysates were prepared. After normalization of the protein content of the cell lysates, luc expression was measured. Data shown represent mean luc expression values ± SDs (calculated from three independent experiments, each of which analysed triplicate wells of a cell culture dish). There was a statistically significant increase in luc expression between the 3JCLI4 (luc) phage particles and the gpD (luc) phage particles, when tested in K562-αvβ3 cells but not when tested in wild-type K562 cells (*P < 0·05, two-way 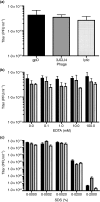

Similar articles
-
A tractable method for simultaneous modifications to the head and tail of bacteriophage lambda and its application to enhancing phage-mediated gene delivery.Nucleic Acids Res. 2007;35(8):e59. doi: 10.1093/nar/gkm146. Epub 2007 Mar 28. Nucleic Acids Res. 2007. PMID: 17392341 Free PMC article.
-
Proteasome inhibitors enhance bacteriophage lambda (lambda) mediated gene transfer in mammalian cells.Virology. 2009 Feb 5;384(1):77-87. doi: 10.1016/j.virol.2008.11.019. Epub 2008 Dec 6. Virology. 2009. PMID: 19064273 Free PMC article.
-
Fc receptor-mediated, antibody-dependent enhancement of bacteriophage lambda-mediated gene transfer in mammalian cells.Virology. 2008 Apr 10;373(2):274-86. doi: 10.1016/j.virol.2007.12.013. Epub 2008 Jan 14. Virology. 2008. PMID: 18191979 Free PMC article.
-
[Vectors--derivatives of phage lambda for construction and analysis of genome libraries].Mol Biol (Mosk). 1987 Sep-Oct;21(5):1172-8. Mol Biol (Mosk). 1987. PMID: 2960881 Review. Russian.
-
Bacteriophage lambda display systems: developments and applications.Appl Microbiol Biotechnol. 2014 Apr;98(7):2853-66. doi: 10.1007/s00253-014-5521-1. Epub 2014 Jan 19. Appl Microbiol Biotechnol. 2014. PMID: 24442507 Review.
Cited by
-
Genetic Engineering and Biosynthesis Technology: Keys to Unlocking the Chains of Phage Therapy.Viruses. 2023 Aug 14;15(8):1736. doi: 10.3390/v15081736. Viruses. 2023. PMID: 37632078 Free PMC article. Review.
-
Engineered Bacteriophage T7 as a Potent Anticancer Agent in vivo.Front Microbiol. 2020 Sep 24;11:491001. doi: 10.3389/fmicb.2020.491001. eCollection 2020. Front Microbiol. 2020. PMID: 33072000 Free PMC article.
-
Recombinant lambda-phage nanobioparticles for tumor therapy in mice models.Genet Vaccines Ther. 2010 May 12;8:3. doi: 10.1186/1479-0556-8-3. Genet Vaccines Ther. 2010. PMID: 20459865 Free PMC article.
-
Communication: Origin of the contributions to DNA structure in phages.J Chem Phys. 2013 Feb 21;138(7):071103. doi: 10.1063/1.4791708. J Chem Phys. 2013. PMID: 23444988 Free PMC article.
-
Phage-based delivery systems: engineering, applications, and challenges in nanomedicines.J Nanobiotechnology. 2024 Jun 25;22(1):365. doi: 10.1186/s12951-024-02576-4. J Nanobiotechnology. 2024. PMID: 38918839 Free PMC article. Review.
References
-
- Amyere M, Mettlen M, Van Der Smissen P, Platek A, Payrastre B, Veithen A, Courtoy PJ. Origin, originality, functions, subversions and molecular signalling of macropinocytosis. Int J Med Microbiol. 2002;291:487–494. - PubMed
-
- Barrow PA, Soothill JS. Bacteriophage therapy and prophylaxis: rediscovery and renewed assessment of potential. Trends Microbiol. 1997;5:268–271. - PubMed
-
- Basner-Tschakarjan E, Mirmohammadsadegh A, Baer A, Hengge UR. Uptake and trafficking of DNA in keratinocytes: evidence for DNA-binding proteins. Gene Ther. 2004;11:765–774. - PubMed
-
- Chen BY, Lim HC. Bioreactor studies on temperature induction of the Q-mutant of bacteriophage lambda in Escherichia coli. J Biotechnol. 1996;51:1–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

