A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data
- PMID: 17448237
- PMCID: PMC1876247
- DOI: 10.1186/1471-2105-8-133
A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data
Abstract
Background: Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) has become one of the most used tools in mass spectrometry based proteomics. Various algorithms have since been developed to automate the process for modern high-throughput LC-MS/MS experiments.
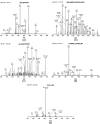
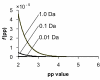
Results: A probability based statistical scoring model for assessing peptide and protein matches in tandem MS database search was derived. The statistical scores in the model represent the probability that a peptide match is a random occurrence based on the number or the total abundance of matched product ions in the experimental spectrum. The model also calculates probability based scores to assess protein matches. Thus the protein scores in the model reflect the significance of protein matches and can be used to differentiate true from random protein matches.
Conclusion: The model is sensitive to high mass accuracy and implicitly takes mass accuracy into account during scoring. High mass accuracy will not only reduce false positives, but also improves the scores of true positive matches. The algorithm is incorporated in an automated database search program MassMatrix.
Figures
References
-
- Nikolaev EN, Somogyi A, Smith DL, Gu CG, Wysocki VH, Martin CD, Samuelson GL. Implementation of low-energy surface-induced dissociation (eV SID) and high-energy collision-induced dissociation (keV CID) in a linear sector-TOF hybrid tandem mass spectrometer. Int J Mass Spectrom. 2001;212:535–551. doi: 10.1016/S1387-3806(01)00462-6. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

