Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene
- PMID: 17449080
- PMCID: PMC2697660
- DOI: 10.1016/j.virol.2007.03.024
Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene
Abstract
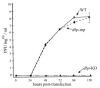
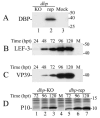
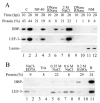
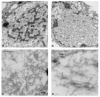
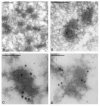
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) encodes two proteins that possess properties typical of single-stranded DNA-binding proteins (SSBs), late expression factor-3 (LEF-3), and a protein referred to as DNA-binding protein (DBP). Whereas LEF-3 is a multi-functional protein essential for viral DNA replication, transporting helicase into the nucleus, and forms a stable complex with the baculovirus alkaline nuclease, the role for DBP in baculovirus replication remains unclear. Therefore, to better understand the functional role of DBP in viral replication, a DBP knockout virus was generated from an AcMNPV bacmid and analyzed. The results of a growth curve analysis indicated that the dbp knockout construct was unable to produce budded virus indicating that dbp is essential. The lack of DBP does not cause a general shutdown of the expression of viral genes, as was revealed by accumulation of early (LEF-3), late (VP39), and very late (P10) proteins in cells transfected with the dbp knockout construct. To investigate the role of DBP in DNA replication, a real-time PCR-based assay was employed and showed that, although viral DNA synthesis occurred in cells transfected with the dbp knockout, the levels were less than that of the control virus suggesting that DBP is required for normal levels of DNA synthesis or for stability of nascent viral DNA. In addition, analysis of the viral DNA replicated by the dbp knockout by using field inversion gel electrophoresis failed to detect the presence of genome-length DNA. Furthermore, analysis of DBP from infected cells indicated that similar to LEF-3, DBP was tightly bound to viral chromatin. Assessment of the cellular localization of DBP relative to replicated viral DNA by immunoelectron microscopy indicated that, at 24 h post-infection, DBP co-localized with nascent DNA at distinct electron-dense regions within the nucleus. Finally, immunoelectron microscopic analysis of cells transfected with the dbp knockout revealed that DBP is required for the production of normal-appearing nucleocapsids and for the generation of the virogenic stroma.
Figures
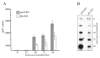

References
-
- Braunagel SC, Parr R, Belyavskyi M, Summers MD. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology. 1998;244:195–211. - PubMed
-
- Busch H. Ubiquitination of proteins. Methods Enzymol. 1984;106:238–62. - PubMed
-
- Campbell MJ. Lipofection reagents prepared by a simple ethanol injection technique. Biotechniques. 1995;18:1027–32. - PubMed
-
- Cunningham RP, Berger H. Mutations affecting genetic recombination in bacteriophage T4D. I. Pathway analysis. Virology. 1977;80:67–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

