DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange
- PMID: 17449621
- PMCID: PMC1913354
- DOI: 10.1128/JB.00376-07
DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange
Abstract
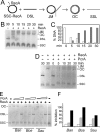
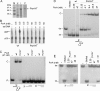
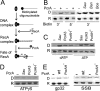
PcrA is a conserved DNA helicase present in all gram-positive bacteria. Bacteria lacking PcrA show high levels of recombination. Lethality induced by PcrA depletion can be overcome by suppressor mutations in the recombination genes recFOR. RecFOR proteins load RecA onto single-stranded DNA during recombination. Here we test whether an essential function of PcrA is to interfere with RecA-mediated DNA recombination in vitro. We demonstrate that PcrA can inhibit the RecA-mediated DNA strand exchange reaction in vitro. Furthermore, PcrA displaced RecA from RecA nucleoprotein filaments. Interestingly, helicase mutants of PcrA also displaced RecA from DNA and inhibited RecA-mediated DNA strand exchange. Employing a novel single-pair fluorescence resonance energy transfer-based assay, we demonstrate a lengthening of double-stranded DNA upon polymerization of RecA and show that PcrA and its helicase mutants can reverse this process. Our results show that the displacement of RecA from DNA by PcrA is not dependent on its translocase activity. Further, our results show that the helicase activity of PcrA, although not essential, might play a facilitatory role in the RecA displacement reaction.
Figures
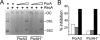

References
-
- Aguilera, A. 2001. Double-strand break repair: are Rad51/RecA-DNA joints barriers to DNA replication? Trends Genet. 17:318-321. - PubMed
-
- Anand, S. P., A. Chattopadhyay, and S. A. Khan. 2005. The PcrA3 mutant binds DNA and interacts with the RepC initiator protein of plasmid pT181 but is defective in its DNA helicase and unwinding activities. Plasmid 54:104-113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

