Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium
- PMID: 17449628
- PMCID: PMC1913349
- DOI: 10.1128/JB.01948-06
Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium
Abstract
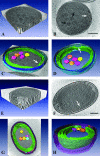
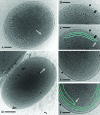
In an age of comparative microbial genomics, knowledge of the near-native architecture of microorganisms is essential for achieving an integrative understanding of physiology and function. We characterized and compared the three-dimensional architecture of the ecologically important cyanobacterium Prochlorococcus in a near-native state using cryo-electron tomography and found that closely related strains have diverged substantially in cellular organization and structure. By visualizing native, hydrated structures within cells, we discovered that the MED4 strain, which possesses one of the smallest genomes (1.66 Mbp) of any known photosynthetic organism, has evolved a comparatively streamlined cellular architecture. This strain possesses a smaller cell volume, an attenuated cell wall, and less extensive intracytoplasmic (photosynthetic) membrane system compared to the more deeply branched MIT9313 strain. Comparative genomic analyses indicate that differences have evolved in key structural genes, including those encoding enzymes involved in cell wall peptidoglycan biosynthesis. Although both strains possess carboxysomes that are polygonal and cluster in the central cytoplasm, the carboxysomes of MED4 are smaller. A streamlined cellular structure could be advantageous to microorganisms thriving in the low-nutrient conditions characteristic of large regions of the open ocean and thus have consequences for ecological niche differentiation. Through cryo-electron tomography we visualized, for the first time, the three-dimensional structure of the extensive network of photosynthetic lamellae within Prochlorococcus and the potential pathways for intracellular and intermembrane movement of molecules. Comparative information on the near-native structure of microorganisms is an important and necessary component of exploring microbial diversity and understanding its consequences for function and ecology.
Figures



References
-
- Badger, M. R., and G. D. Price. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609-622. - PubMed
-
- Bergthorsson, U., and H. Ochman. 1998. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 15:9-16. - PubMed
-
- Campbell, L., H. B. Liu, H. A. Nolla, and D. Vaulot. 1997. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991-1994 ENSO event. Deep-Sea Res. Part I 44:167.
-
- Crowther, R. A., D. J. DeRosier, and A. Klug. 1970. The reconstruction of a three-dimensional structure from its projections and its application to electron microscopy. Proc. R. Soc. Lond. A 317:319-340.
-
- Dmitriev, B., F. Toukach, and S. Ehlers. 2005. Towards a comprehensive view of the bacterial cell wall. Trends Microbiol. 13:569-574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

