More genes underwent positive selection in chimpanzee evolution than in human evolution
- PMID: 17449636
- PMCID: PMC1863478
- DOI: 10.1073/pnas.0701705104
More genes underwent positive selection in chimpanzee evolution than in human evolution
Abstract
Observations of numerous dramatic and presumably adaptive phenotypic modifications during human evolution prompt the common belief that more genes have undergone positive Darwinian selection in the human lineage than in the chimpanzee lineage since their evolutionary divergence 6-7 million years ago. Here, we test this hypothesis by analyzing nearly 14,000 genes of humans and chimps. To ensure an accurate and unbiased comparison, we select a proper outgroup, avoid sequencing errors, and verify statistical methods. Our results show that the number of positively selected genes is substantially smaller in humans than in chimps, despite a generally higher nonsynonymous substitution rate in humans. These observations are explainable by the reduced efficacy of natural selection in humans because of their smaller long-term effective population size but refute the anthropocentric view that a grand enhancement in Darwinian selection underlies human origins. Although human and chimp positively selected genes have different molecular functions and participate in different biological processes, the differences do not ostensibly correspond to the widely assumed adaptations of these species, suggesting how little is currently known about which traits have been under positive selection. Our analysis of the identified positively selected genes lends support to the association between human Mendelian diseases and past adaptations but provides no evidence for either the chromosomal speciation hypothesis or the widespread brain-gene acceleration hypothesis of human origins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

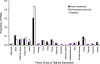
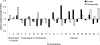
References
-
- Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

