Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria
- PMID: 17449644
- PMCID: PMC1914192
- DOI: 10.1104/pp.107.095414
Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria
Abstract
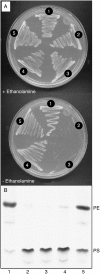
Phosphatidylserine (PS) decarboxylase is involved in the synthesis of the abundant phospholipid phosphatidylethanolamine (PE), particularly in mitochondria, in many organisms, including yeast (Saccharomyces cerevisiae) and animals. Arabidopsis (Arabidopsis thaliana) contains three genes with sequence similarity to PS decarboxylases, and the respective gene products were functionally characterized after heterologous expression in yeast and Escherichia coli. While the PSD1 protein localizes to mitochondria, PSD2 and PSD3 are found in the endomembrane system. To study the role of PSD genes in plant phospholipid metabolism, Arabidopsis insertional mutants for psd1, psd2, and psd3 were obtained. The single mutants were decreased in PS decarboxylase activity to various extents, but mutant plants showed no obvious growth or morphological phenotype. A triple mutant, psd1 psd2 psd3, was generated that was totally devoid of PS decarboxylase activity. While the phospholipid composition in whole leaves was unchanged, the PE content in isolated mitochondria of psd1 psd2 psd3 was decreased. Therefore, the predominant proportion of PE in Arabidopsis is synthesized by alternative pathways, but a significant amount of mitochondrial PE is derived from the PS decarboxylase reaction. These results imply that, similar to yeast and animal cells, a specific phospholipid transfer from the endoplasmic reticulum to mitochondria exists in plants.
Figures





Similar articles
-
Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae.J Biol Chem. 1995 Mar 17;270(11):6062-70. doi: 10.1074/jbc.270.11.6062. J Biol Chem. 1995. PMID: 7890739
-
Mitochondrial phosphatidylserine decarboxylase from higher plants. Functional complementation in yeast, localization in plants, and overexpression in Arabidopsis.Plant Physiol. 2003 Jul;132(3):1678-87. doi: 10.1104/pp.103.023242. Plant Physiol. 2003. PMID: 12857846 Free PMC article.
-
Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiáe. Cloning and mapping of the gene, heterologous expression, and creation of the null allele.J Biol Chem. 1995 Mar 17;270(11):6071-80. doi: 10.1074/jbc.270.11.6071. J Biol Chem. 1995. PMID: 7890740
-
Cell biology, physiology and enzymology of phosphatidylserine decarboxylase.Biochim Biophys Acta Mol Cell Biol Lipids. 2017 Jan;1862(1):25-38. doi: 10.1016/j.bbalip.2016.09.007. Epub 2016 Sep 17. Biochim Biophys Acta Mol Cell Biol Lipids. 2017. PMID: 27650064 Review.
-
Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells.Biochim Biophys Acta. 2013 Mar;1831(3):543-54. doi: 10.1016/j.bbalip.2012.08.016. Epub 2012 Aug 29. Biochim Biophys Acta. 2013. PMID: 22960354 Review.
Cited by
-
Mitochondrial fatty acid synthesis is required for normal mitochondrial morphology and function in Trypanosoma brucei.Mol Microbiol. 2008 Mar;67(5):1125-42. doi: 10.1111/j.1365-2958.2008.06112.x. Epub 2008 Jan 23. Mol Microbiol. 2008. PMID: 18221265 Free PMC article.
-
Glycerolipid synthesis and lipid trafficking in plant mitochondria.FEBS J. 2017 Feb;284(3):376-390. doi: 10.1111/febs.13812. Epub 2016 Aug 1. FEBS J. 2017. PMID: 27406373 Free PMC article. Review.
-
Role of aminoalcoholphosphotransferases 1 and 2 in phospholipid homeostasis in Arabidopsis.Plant Cell. 2015 May;27(5):1512-28. doi: 10.1105/tpc.15.00180. Epub 2015 May 5. Plant Cell. 2015. PMID: 25944098 Free PMC article.
-
Acyl-lipid metabolism.Arabidopsis Book. 2010;8:e0133. doi: 10.1199/tab.0133. Epub 2010 Jun 11. Arabidopsis Book. 2010. PMID: 22303259 Free PMC article.
-
Compartment-specific synthesis of phosphatidylethanolamine is required for normal heavy metal resistance.Mol Biol Cell. 2010 Feb 1;21(3):443-55. doi: 10.1091/mbc.e09-06-0519. Epub 2009 Dec 16. Mol Biol Cell. 2010. PMID: 20016005 Free PMC article.
References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Amann E, Brosius J (1985) ATG vectors for regulated high-level expression of cloned genes in Escherichia coli. Gene 40 183–190 - PubMed
-
- Beisson F, Koo AJK, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al (2003) Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol 132 681–697 - PMC - PubMed
-
- Benning C, Huang Y-H, Gage DA (1995) Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys 317 103–111 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

