A comprehensive calorimetric investigation of an entropically driven T cell receptor-peptide/major histocompatibility complex interaction
- PMID: 17449678
- PMCID: PMC1896243
- DOI: 10.1529/biophysj.107.104570
A comprehensive calorimetric investigation of an entropically driven T cell receptor-peptide/major histocompatibility complex interaction
Abstract
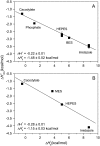
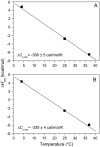
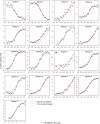
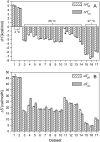
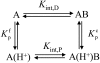
The alphabeta T cell receptor (TCR) is responsible for recognizing peptides bound and "presented" by major histocompatibility complex (MHC) molecules. We recently reported that at 25 degrees C the A6 TCR, which recognizes the Tax peptide presented by the class I MHC human leukocyte antigen-A*0201 (HLA-A2), binds with a weak DeltaH degrees , a favorable DeltaS degrees , and a moderately negative DeltaC(p). These observations were of interest given the unfavorable binding entropies and large heat capacity changes measured for many other TCR-ligand interactions, suggested to result from TCR conformational changes occurring upon binding. Here, we further investigated the A6-Tax/HLA-A2 interaction using titration calorimetry. We found that binding results in a pK(a) shift, complicating interpretation of measured binding thermodynamics. To better characterize the interaction, we measured binding as a function of pH, temperature, and buffer ionization enthalpy. A global analysis of the resulting data allowed determination of both the intrinsic binding thermodynamics separated from the influence of protonation as well as the thermodynamics associated with the pK(a) shift. Our results indicate that intrinsically, A6 binds Tax/HLA-A2 with a very weak DeltaH degrees , an even more favorable DeltaS degrees than previously thought, and a relatively large negative DeltaC(p). Comparison of these energetics with the makeup of the protein-protein interface suggests that conformational adjustments are required for binding, but these are more likely to be structural shifts, rather than disorder-to-order transitions. The thermodynamics of the pK(a) shift suggest protonation may be linked to an additional process such as ion binding.
Figures



References
-
- Rudolph, M. G., R. L. Stanfield, and I. A. Wilson. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466. - PubMed
-
- Davis, M. M., J. J. Boniface, Z. Reich, D. Lyons, J. Hampl, B. Arden, and Y. Chien. 1998. Ligand recognition by αβ T cell receptors. Annu. Rev. Immunol. 16:523–544. - PubMed
-
- Willcox, B. E., G. F. Gao, J. R. Wyer, J. E. Ladbury, J. I. Bell, B. K. Jakobsen, and P. A. van der Merwe. 1999. TCR binding to peptide-MHC stabilizes a flexible recognition interface. Immunity. 10:357–365. - PubMed
-
- Anikeeva, N., T. Lebedeva, M. Krogsgaard, S. Y. Tetin, E. Martinez-Hackert, S. A. Kalams, M. M. Davis, and Y. Sykulev. 2003. Distinct molecular mechanisms account for the specificity of two different T-cell receptors. Biochemistry. 42:4709–4716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

