Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization
- PMID: 17449691
- PMCID: PMC1932723
- DOI: 10.1128/AEM.00147-07
Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization
Abstract
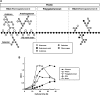
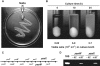
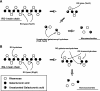
Plant cell wall degradation is a premier event when Bacillus subtilis, a typical saprophytic bacterium, invades plants. Here we show the degradation system of rhamnogalacturonan type I (RG-I), a component of pectin from the plant cell wall, in B. subtilis strain 168. Strain 168 cells showed a significant growth on plant cell wall polysaccharides such as pectin, polygalacturonan, and RG-I as a carbon source. DNA microarray analysis indicated that three gene clusters (yesOPQRSTUVWXYZ, ytePQRST, and ybcMOPST-ybdABDE) are inducibly expressed in strain 168 cells grown on RG-I. Cells of an industrially important bacterium, B. subtilis strain natto, fermenting soybeans also express the gene cluster including the yes series during the assimilation of soybean used as a carbon source. Among proteins encoded in the yes cluster, YesW and YesX were found to be novel types of RG lyases releasing disaccharide from RG-I. Genetic and enzymatic properties of YesW and YesX suggest that strain 168 cells secrete YesW, which catalyzes the initial cleavage of the RG-I main chain, and the resultant oligosaccharides are converted to disaccharides through the extracellular exotype YesX reaction. The disaccharide is finally degraded into its constituent monosaccharides through the reaction of intracellular unsaturated galacturonyl hydrolases YesR and YteR. This enzymatic route for RG-I degradation in strain 168 differs significantly from that in plant-pathogenic fungus Aspergillus aculeatus. This is, to our knowledge, the first report on the bacterial system for complete RG-I main chain degradation.
Figures


References
-
- Akiyama, H., T. Endo, R. Nakakita, K. Murata, Y. Yonemoto, and K. Okayama. 1992. Effect of depolymerized alginates on the growth of bifidobacteria. Biosci. Biotechnol. Biochem. 56:355-356. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Carpita, N. C., and D. M. Gibeaut. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3:1-30. - PubMed
-
- Clements, L. D., U. N. Streips, and B. S. Miller. 2002. Differential proteomic analysis of Bacillus subtilis nitrate respiration and fermentation in defined medium. Proteomics 2:1724-1734. - PubMed
-
- Darvill, A., C. Bergmann, F. Cervone, G. De Lorenzo, K. S. Ham, M. D. Spiro, W. S. York, and P. Albersheim. 1994. Oligosaccharins involved in plant growth and host-pathogen interactions. Biochem. Soc. Symp. 60:89-94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

