Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains
- PMID: 17449699
- PMCID: PMC1932674
- DOI: 10.1128/AEM.02735-06
Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains
Abstract
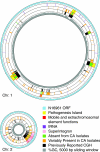
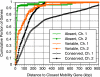
Vibrio cholerae is an autochthonous member of diverse aquatic ecosystems around the globe. Collectively, the genomes of environmental V. cholerae strains comprise a large repository of encoded functions which can be acquired by individual V. cholerae lineages through uptake and recombination. To characterize the genomic diversity of environmental V. cholerae, we used comparative genome hybridization to study 41 environmental strains isolated from diverse habitats along the central California coast, a region free of endemic cholera. These data were used to classify genes of the epidemic V. cholerae O1 sequenced strain N16961 as conserved, variably present, or absent from the isolates. For the most part, absent genes were restricted to large mobile elements and have known functions in pathogenesis. Conversely, genes present in some, but not all, California isolates were in smaller contiguous clusters and were less likely to be near genes with functions in DNA mobility. Two such clusters of variable genes encoding different selectable metabolic phenotypes (mannose and diglucosamine utilization) were transformed into the genomes of environmental isolates by chitin-dependent competence, indicating that this mechanism of general genetic exchange is conserved among V. cholerae. The transformed DNA had an average size of 22.7 kbp, demonstrating that natural competence can mediate the movement of large chromosome fragments. Thus, whether variable genes arise through the acquisition of new sequences by horizontal gene transfer or by the loss of preexisting DNA though deletion, natural transformation provides a mechanism by which V. cholerae clones can gain access to the V. cholerae pan-genome.
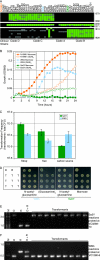
Figures

References
-
- Ansaruzzaman, M., M. J. Albert, I. Kuhn, S. M. Faruque, A. K. Siddique, and R. Mollby. 1996. Differentiation of Vibrio cholerae O1 isolates with biochemical fingerprinting and comparison with ribotyping. J. Diarrhoeal Dis. Res. 14:248-254. - PubMed
-
- Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Short protocols in molecular biology. Wiley, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

