A family of poly(U) polymerases
- PMID: 17449726
- PMCID: PMC1869031
- DOI: 10.1261/rna.514007
A family of poly(U) polymerases
Abstract
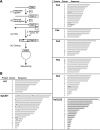
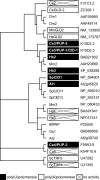
The GLD-2 family of poly(A) polymerases add successive AMP monomers to the 3' end of specific RNAs, forming a poly(A) tail. Here, we identify a new group of GLD-2-related nucleotidyl transferases from Arabidopsis, Schizosaccharomyces pombe, Caenorhabditis elegans, and humans. Like GLD-2, these enzymes are template independent and add nucleotides to the 3' end of an RNA substrate. However, these new enzymes, which we refer to as poly(U) polymerases, add poly(U) rather than poly(A) to their RNA substrates.
Figures




References
-
- Aphasizhev, R., Sbicego, S., Peris, M., Jang, S.H., Aphasizheva, I., Simpson, A.M., Rivlin, A., Simpson, L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. - PubMed
-
- Barnard, D.C., Ryan, K., Manley, J.L., Richter, J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
