An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction
- PMID: 17449807
- PMCID: PMC1913751
- DOI: 10.1105/tpc.106.048264
An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction
Abstract
Rhizobial bacteria activate the formation of nodules on the appropriate host legume plant, and this requires the bacterial signaling molecule Nod factor. Perception of Nod factor in the plant leads to the activation of a number of rhizobial-induced genes. Putative transcriptional regulators in the GRAS family are known to function in Nod factor signaling, but these proteins have not been shown to be capable of direct DNA binding. Here, we identify an ERF transcription factor, ERF Required for Nodulation (ERN), which contains a highly conserved AP2 DNA binding domain, that is necessary for nodulation. Mutations in this gene block the initiation and development of rhizobial invasion structures, termed infection threads, and thus block nodule invasion by the bacteria. We show that ERN is necessary for Nod factor-induced gene expression and for spontaneous nodulation activated by the calcium- and calmodulin-dependent protein kinase, DMI3, which is a component of the Nod factor signaling pathway. We propose that ERN is a component of the Nod factor signal transduction pathway and functions downstream of DMI3 to activate nodulation gene expression.
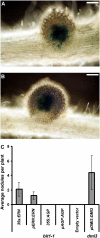
Figures






References
-
- Ane, J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. - PubMed
-
- Ardourel, M., Demont, N., Debelle, F.D., Maillet, F., Debilly, F., Prome, J.C., Denarie, J., and Truchet, G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

