MRI in mouse developmental biology
- PMID: 17451170
- PMCID: PMC2694493
- DOI: 10.1002/nbm.1146
MRI in mouse developmental biology
Abstract
Mice are used in many studies to determine the role of genetic and molecular factors in mammalian development and human congenital diseases. MRI has emerged as a major method for analyzing mutant and transgenic phenotypes in developing mice, at both embryonic and neonatal stages. Progress in this area is reviewed, with emphasis on the use of MRI to analyze cardiovascular and neural development in mice. Comparisons are made with other imaging technologies, including optical and ultrasound imaging, discussing the potential strengths and weaknesses of MRI and identifying the future challenges for MRI in mouse developmental biology.
Copyright (c) 2007 John Wiley & Sons, Ltd.
Figures
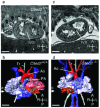



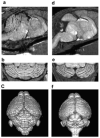
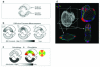

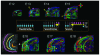
References
-
- Smith BR, Linney E, Huff DS, Johnson GA. Magnetic resonance microscopy of embryos. Comput Med Imaging Graph. 1996;20:483–490. - PubMed
-
- Smith BR. Magnetic resonance microscopy in cardiac development [Review] Microsc Res Tech. 2001;52:323–330. - PubMed
-
- Huang GY, Wessels A, Smith BR, Linask KK, Ewart JL, Lo CW. Alteration in connexin 43 gap junction gene dosage impairs conotruncal heart development. Dev Biol. 1998;198:32–44. - PubMed
-
- Dhenain M, Ruffins SW, Jacobs RE. Three-dimensional digital mouse atlas using high-resolution MRI. Dev Biol. 2001;232:458–470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

