Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system
- PMID: 17451448
- PMCID: PMC1974823
- DOI: 10.1111/j.1574-6968.2007.00722.x
Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system
Abstract
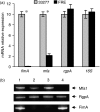
Porphyromonas gingivalis possesses two distinct fimbriae. The long (FimA) fimbriae have been extensively studied. Expression of the fimA gene is tightly controlled by a two-component system (FimS/FimR) through a cascade regulation. The short (Mfa1) fimbriae are less understood. The authors have recently demonstrated that both fimbriae are required for formation of P. gingivalis biofilms. Here, the novel finding that FimR, a member of the two-component regulatory system, is a transcriptional activator of the mfa1 gene is promoted. Unlike the regulatory mechanism of FimA by FimR, this regulation of the mfa1 gene is accomplished by FimR directly binding to the promoter region of mfa1.
Figures


References
-
- Amano A, Nakagawa I, Okahashi N, Hamada N. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res. 2004;39:136–142. - PubMed
-
- Boggess KA, Madianos PN, Preisser JS, Moise KJ, Jr, Offenbacher S. Chronic maternal and fetal Porphyromonas gingivalis exposure during pregnancy in rabbits. Am J Obstet Gynecol. 2005;192:554–557. - PubMed
-
- Brodala N, Merricks EP, Bellinger DA, et al. Porphyromonas gingivalis bacteremia induces coronary and aortic atherosclerosis in normocholesterolemic and hypercholesterolemic pigs. Arterioscler Thromb Vasc Biol. 2005;25:1446–1451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

