hnRNP E1 and E2 have distinct roles in modulating HIV-1 gene expression
- PMID: 17451601
- PMCID: PMC1863430
- DOI: 10.1186/1742-4690-4-28
hnRNP E1 and E2 have distinct roles in modulating HIV-1 gene expression
Abstract
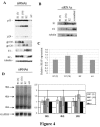
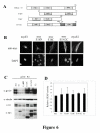
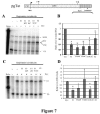
Pre-mRNA processing, including 5' end capping, splicing, and 3' end cleavage/polyadenylation, are events coordinated by transcription that can influence the subsequent export and translation of mRNAs. Coordination of RNA processing is crucial in retroviruses such as HIV-1, where inefficient splicing and the export of intron-containing RNAs are required for expression of the full complement of viral proteins. RNA processing can be affected by both viral and cellular proteins, and in this study we demonstrate that a member of the hnRNP E family of proteins can modulate HIV-1 RNA metabolism and expression. We show that hnRNP E1/E2 are able to interact with the ESS3a element of the bipartite ESS in tat/rev exon 3 of HIV-1 and that modulation of hnRNP E1 expression alters HIV-1 structural protein synthesis. Overexpression of hnRNP E1 leads to a reduction in Rev, achieved in part through a decrease in rev mRNA levels. However, the reduction in Rev levels cannot fully account for the effect of hnRNP E1, suggesting that hmRNP E1 might also act to suppress viral RNA translation. Deletion mutagenesis determined that the C-terminal end of hnRNP E1 was required for the reduction in Rev expression and that replacing this portion of hnRNP E1 with that of hnRNP E2, despite the high degree of conservation, could not rescue the loss of function.
Figures





References
-
- Dirks RW, Hattinger CM, Molenaar C, Snaar SP. Synthesis, processing, and transport of RNA within the three-dimensional context of the cell nucleus. Critical Reviews in Eukaryotic Gene Expression. 1999;9:191–201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

