Silencing and complementation of reovirus core protein mu2: functional correlations with mu2-microtubule association and differences between virus- and plasmid-derived mu2
- PMID: 17451769
- PMCID: PMC2486448
- DOI: 10.1016/j.virol.2007.03.037
Silencing and complementation of reovirus core protein mu2: functional correlations with mu2-microtubule association and differences between virus- and plasmid-derived mu2
Abstract
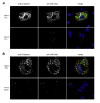
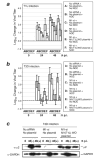
A low-copy component of mammalian reovirus particles is mu2, an 83-kDa protein encoded by the M1 viral genome segment and packaged within the viral core. Previous studies have identified mu2 as a nucleoside triphosphate phosphohydrolase (NTPase) as well as an RNA 5'-triphosphate phosphohydrolase (RTPase), putatively involved in reovirus RNA synthesis and/or 5'-capping. Other studies have identified mu2 as a microtubule-binding protein, which also associates with the viral factory matrix protein muNS and thereby anchors the factories to cellular microtubules during infections by most reovirus strains. To extend studies of mu2 functions during infection, we tested a small interfering RNA (siRNA) directed against the M1 plus-strand RNAs of reovirus strains Type 1 Lang (T1L) and Type 3 Dearing (T3D). The siRNA strongly suppressed mu2 expression by either strain and reduced infectious yields in a strain-dependent manner. This first strain difference was genetically mapped to the M1 genome segment and tentatively assigned to a single mu2 sequence polymorphism, Pro/Ser208, which also determines a T1L-T3D strain difference in microtubule association. The siRNA-based defect in mu2 expression was rescued by plasmids, containing silent mutations in the siRNA-targeted sequence, which encoded either T1L or T3D mu2, but the growth defect was rescued only by T1L mu2. This second strain difference was also mapped to Pro/Ser208, in that swapping this one residue between T1L and T3D mu2 reversed the rescue phenotypes. Thus, the T1L-T3D strain difference in mu2-microtubule association was correlated not only with the extent of reduction in infectious yields by the siRNA but also with the extent of rescue by plasmid-derived mu2. In addition, the rescue capacity of T1L mu2 was abrogated by nocodazole treatment, providing independent evidence for the importance of mu2-microtubule association in plasmid-based rescue. In two separate cases, the results revealed functional differences between virus- and plasmid-derived mu2. Ala substitutions within the NTP-binding motif of T1L mu2 also abrogated its rescue capacity, suggesting that the NTPase or RTPase activity of mu2 is additionally required for effective viral growth.
Figures






Similar articles
-
Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules.J Virol. 2002 May;76(9):4483-96. doi: 10.1128/jvi.76.9.4483-4496.2002. J Virol. 2002. PMID: 11932414 Free PMC article.
-
Comparisons of the M1 genome segments and encoded mu2 proteins of different reovirus isolates.Virol J. 2004 Sep 23;1:6. doi: 10.1186/1743-422X-1-6. Virol J. 2004. PMID: 15507160 Free PMC article.
-
Gene-specific inhibition of reovirus replication by RNA interference.J Virol. 2006 Sep;80(18):9053-63. doi: 10.1128/JVI.00276-06. J Virol. 2006. PMID: 16940517 Free PMC article.
-
The M2 Gene Is a Determinant of Reovirus-Induced Myocarditis.J Virol. 2022 Jan 26;96(2):e0187921. doi: 10.1128/JVI.01879-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757847 Free PMC article.
-
The influence of the cytoskeleton on the development and behavior of viral factories in mammalian orthoreovirus.Virology. 2025 Mar;604:110423. doi: 10.1016/j.virol.2025.110423. Epub 2025 Jan 27. Virology. 2025. PMID: 39889480 Review.
Cited by
-
Sequestration of free tubulin molecules by the viral protein NSP2 induces microtubule depolymerization during rotavirus infection.J Virol. 2010 Mar;84(5):2522-32. doi: 10.1128/JVI.01883-09. Epub 2009 Dec 23. J Virol. 2010. PMID: 20032187 Free PMC article.
-
Dissection of mammalian orthoreovirus µ2 reveals a self-associative domain required for binding to microtubules but not to factory matrix protein µNS.PLoS One. 2017 Sep 7;12(9):e0184356. doi: 10.1371/journal.pone.0184356. eCollection 2017. PLoS One. 2017. PMID: 28880890 Free PMC article.
-
Reovirus mu2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9.J Virol. 2009 Mar;83(5):2178-87. doi: 10.1128/JVI.01787-08. Epub 2008 Dec 24. J Virol. 2009. PMID: 19109390 Free PMC article.
-
Engineering recombinant reoviruses with tandem repeats and a tetravirus 2A-like element for exogenous polypeptide expression.Proc Natl Acad Sci U S A. 2013 May 14;110(20):E1867-76. doi: 10.1073/pnas.1220107110. Epub 2013 Apr 29. Proc Natl Acad Sci U S A. 2013. PMID: 23630248 Free PMC article.
-
A post-entry step in the mammalian orthoreovirus replication cycle is a determinant of cell tropism.J Biol Chem. 2010 Dec 31;285(53):41604-13. doi: 10.1074/jbc.M110.176255. Epub 2010 Oct 26. J Biol Chem. 2010. PMID: 20978124 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

