OligoCalc: an online oligonucleotide properties calculator
- PMID: 17452344
- PMCID: PMC1933198
- DOI: 10.1093/nar/gkm234
OligoCalc: an online oligonucleotide properties calculator
Abstract
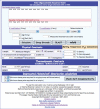
We developed OligoCalc as a web-accessible, client-based computational engine for reporting DNA and RNA single-stranded and double-stranded properties, including molecular weight, solution concentration, melting temperature, estimated absorbance coefficients, inter-molecular self-complementarity estimation and intra-molecular hairpin loop formation. OligoCalc has a familiar 'calculator' look and feel, making it readily understandable and usable. OligoCalc incorporates three common methods for calculating oligonucleotide-melting temperatures, including a nearest-neighbor thermodynamic model for melting temperature. Since it first came online in 1997, there have been more than 900,000 accesses of OligoCalc from nearly 200,000 distinct hosts, excluding search engines. OligoCalc is available at http://basic.northwestern.edu/biotools/OligoCalc.html, with links to the full source code, usage patterns and statistics at that link as well.
Figures
References
-
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. - PubMed
-
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001.
-
- Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 1962;5:109–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources