Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes
- PMID: 17452368
- PMCID: PMC1888817
- DOI: 10.1093/nar/gkm111
Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes
Abstract
Parallel tetramolecular quadruplexes may be formed with short oligodeoxynucleotides bearing a block of three or more guanines. We analyze the properties of sequence variants of parallel quadruplexes in which each guanine of the central block was systematically substituted with a different base. Twelve types of substitutions were assessed in more than 100 different sequences. We conducted a comparative kinetic analysis of all tetramers. Electrospray mass spectrometry was used to count the number of inner cations, which is an indicator of the number of effective tetrads. In general, the presence of a single substitution has a strong deleterious impact on quadruplex stability, resulting in reduced quadruplex lifetime/thermal stability and in decreased association rate constants. We demonstrate extremely large differences in the association rate constants of these quadruplexes depending on modification position and type. These results demonstrate that most guanine substitutions are deleterious to tetramolecular quadruplex structure. Despite the presence of well-defined non-guanine base quartets in a number of NMR and X-ray structures, our data suggest that most non-guanine quartets do not participate favorably in structural stability, and that these quartets are formed only by virtue of the docking platform provided by neighboring G-quartets. Two notable exceptions were found with 8-bromo-guanine (X) and 6-methyl-isoxanthopterin (P) substitutions, which accelerate quadruplex formation by a factor of 10 when present at the 5' end. The thermodynamic and kinetic data compiled here are highly valuable for the design of DNA quadruplex assemblies with tunable association/dissociation properties.
Figures
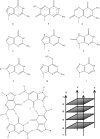
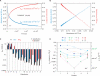
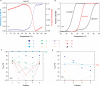
 (red). ‘-’ corresponds to TG4T. Note that these relative values have been normalized for each cation compared to the unmodified oligonucleotide. kon values in K+ and Na+ are respectively 2000 and 10 times higher than in
(red). ‘-’ corresponds to TG4T. Note that these relative values have been normalized for each cation compared to the unmodified oligonucleotide. kon values in K+ and Na+ are respectively 2000 and 10 times higher than in  . (D) Association constants in Na+: Effect of a single guanine substitution on kon (corresponding curves in K+ or
. (D) Association constants in Na+: Effect of a single guanine substitution on kon (corresponding curves in K+ or  are provided as Supplementary Data). The position of the substitution is indicated on the X-axis: position 1 corresponds to the first guanine (5′ side), position 5 to the last guanine (3′ side). The relative kon values (±SD) for the formation of the TG5T variants are indicated on the left Y-axis (kon for the unmodified TG5T sequence under the same conditions = 1, corresponding to a horizontal dotted line). Absolute values are shown on the right Y-axis. Experiments were performed in 0.11 M Na+ at 3 ± 1°C. Note the semi-log scale: for many mutants, a single substitution may lead a tremendous decrease in kon. Only a few cases lead to a higher kon than TG5T, for example T
are provided as Supplementary Data). The position of the substitution is indicated on the X-axis: position 1 corresponds to the first guanine (5′ side), position 5 to the last guanine (3′ side). The relative kon values (±SD) for the formation of the TG5T variants are indicated on the left Y-axis (kon for the unmodified TG5T sequence under the same conditions = 1, corresponding to a horizontal dotted line). Absolute values are shown on the right Y-axis. Experiments were performed in 0.11 M Na+ at 3 ± 1°C. Note the semi-log scale: for many mutants, a single substitution may lead a tremendous decrease in kon. Only a few cases lead to a higher kon than TG5T, for example T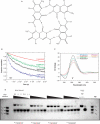

 ions present in the quadruplexes: MS analysis. ESI-MS spectra obtained in gentle condition help in understanding the formation of these tetramolecular structures, not only by providing the strand stoichiometry but also an unambiguous determination of the number of contributing structural cations. The position of each substitution is indicated on the X-axis. The mean number of ammonium ions (NNH4) present in the complexes is obtained from equation: NNH4 = [4 × I(G4with 4NH4) + 3 × I(G4with 3NH4) + 2 × I(G4with 2NH4) + 1 × I(G4with 1NH4)]/Sum[I(G4all)] where I(G4n) are the relative intensities of the quadruplex with different number of ammonium ions. Note that the
ions present in the quadruplexes: MS analysis. ESI-MS spectra obtained in gentle condition help in understanding the formation of these tetramolecular structures, not only by providing the strand stoichiometry but also an unambiguous determination of the number of contributing structural cations. The position of each substitution is indicated on the X-axis. The mean number of ammonium ions (NNH4) present in the complexes is obtained from equation: NNH4 = [4 × I(G4with 4NH4) + 3 × I(G4with 3NH4) + 2 × I(G4with 2NH4) + 1 × I(G4with 1NH4)]/Sum[I(G4all)] where I(G4n) are the relative intensities of the quadruplex with different number of ammonium ions. Note that the Similar articles
-
Intramolecular and intermolecular guanine quadruplexes of DNA in aqueous salt and ethanol solutions.Biopolymers. 2007 May;86(1):1-10. doi: 10.1002/bip.20672. Biopolymers. 2007. PMID: 17211886
-
Kinetics of double-chain reversals bridging contiguous quartets in tetramolecular quadruplexes.Nucleic Acids Res. 2006 May 8;34(8):2386-97. doi: 10.1093/nar/gkl098. Print 2006. Nucleic Acids Res. 2006. PMID: 16682446 Free PMC article.
-
Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements.Biochemistry. 2004 Apr 27;43(16):4877-84. doi: 10.1021/bi0300985. Biochemistry. 2004. PMID: 15096057
-
Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids.Biopolymers. 2000-2001;56(3):147-94. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. Biopolymers. 2000. PMID: 11745110 Review.
-
Quadruplex structures in nucleic acids.Biopolymers. 2000-2001;56(3):123-46. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. Biopolymers. 2000. PMID: 11745109 Review.
Cited by
-
Synthesis and g-quadruplex-binding properties of defined acridine oligomers.J Nucleic Acids. 2010 Jun 13;2010:489060. doi: 10.4061/2010/489060. J Nucleic Acids. 2010. PMID: 20725626 Free PMC article.
-
Highly fluorescent guanosine mimics for folding and energy transfer studies.Nucleic Acids Res. 2011 Aug;39(15):6825-34. doi: 10.1093/nar/gkr281. Epub 2011 May 6. Nucleic Acids Res. 2011. PMID: 21551219 Free PMC article.
-
Xanthine and 8-oxoguanine in G-quadruplexes: formation of a G·G·X·O tetrad.Nucleic Acids Res. 2015 Dec 2;43(21):10506-14. doi: 10.1093/nar/gkv826. Epub 2015 Sep 22. Nucleic Acids Res. 2015. PMID: 26400177 Free PMC article.
-
Mixed guanine, adenine base quartets: possible roles of protons and metal ions in their stabilization.J Biol Inorg Chem. 2018 Jan;23(1):41-49. doi: 10.1007/s00775-017-1507-7. Epub 2017 Dec 7. J Biol Inorg Chem. 2018. PMID: 29218641 Free PMC article.
-
Ball with hair: modular functionalization of highly stable G-quadruplex DNA nano-scaffolds through N2-guanine modification.Nucleic Acids Res. 2017 Jun 20;45(11):6265-6274. doi: 10.1093/nar/gkx243. Nucleic Acids Res. 2017. PMID: 28499037 Free PMC article.
References
-
- Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

