Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia
- PMID: 17456006
- PMCID: PMC1854911
- DOI: 10.1371/journal.pbio.0050118
Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia
Abstract
Despite its morphological similarity to the other species in the Drosophila melanogaster species complex, D. sechellia has evolved distinct physiological and behavioral adaptations to its host plant Morinda citrifolia, commonly known as Tahitian Noni. The odor of the ripe fruit of M. citrifolia originates from hexanoic and octanoic acid. D. sechellia is attracted to these two fatty acids, whereas the other species in the complex are repelled. Here, using interspecies hybrids between D. melanogaster deficiency mutants and D. sechellia, we showed that the Odorant-binding protein 57e (Obp57e) gene is involved in the behavioral difference between the species. D. melanogaster knock-out flies for Obp57e and Obp57d showed altered behavioral responses to hexanoic acid and octanoic acid. Furthermore, the introduction of Obp57d and Obp57e from D. simulans and D. sechellia shifted the oviposition site preference of D. melanogaster Obp57d/e(KO) flies to that of the original species, confirming the contribution of these genes to D. sechellia's specialization to M. citrifolia. Our finding of the genes involved in host-plant determination may lead to further understanding of mechanisms underlying taste perception, evolution of plant-herbivore interactions, and speciation.
Conflict of interest statement
Figures



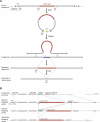

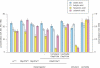
Comment in
-
Can a taste for poison drive speciation?PLoS Biol. 2007 May;5(5):e140. doi: 10.1371/journal.pbio.0050140. Epub 2007 Apr 24. PLoS Biol. 2007. PMID: 20076673 Free PMC article. No abstract available.
References
-
- Bernays EA, Chapman RF. Host-plant selection by phytophagous insects. New York: Chapman & Hall; 1994. 312
-
- Strauss SY, Rudgers JA, Lau JA, Irwin R. Direct and ecological costs of resistance to herbivory. Trends Ecol Evol. 2002;17:278–285.
-
- Funk DJ, Filchak KE, Feder JL. Herbivorous insects: Model systems for the comparative study of speciation ecology. Genetica. 2002;116:251–267. - PubMed
-
- Mallet J. What does Drosophila genetics tell us about speciation? Trends Ecol Evol. 2006;21:386–393. - PubMed
-
- Chapman RF. Contact chemoreception in feeding by phytophagous insects. Ann Rev Entmol. 2003;48:455–484. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

