Discovery of tissue-specific exons using comprehensive human exon microarrays
- PMID: 17456239
- PMCID: PMC1896007
- DOI: 10.1186/gb-2007-8-4-r64
Discovery of tissue-specific exons using comprehensive human exon microarrays
Abstract
Background: Higher eukaryotes express a diverse population of messenger RNAs generated by alternative splicing. Large-scale methods for monitoring gene expression must adapt in order to accurately detect the transcript variation generated by this splicing.
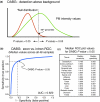
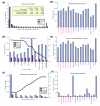
Results: We have designed a high-density oligonucleotide microarray with probesets for more than one million annotated and predicted exons in the human genome. Using these arrays and a simple algorithm that normalizes exon signal to signal from the gene as a whole, we have identified tissue-specific exons from a panel of 16 different normal adult tissues. RT-PCR validation confirms approximately 86% of the predicted tissue-enriched probesets. Pair-wise comparisons between the tissues suggest that as many as 73% of detected genes are differentially alternatively spliced. We also demonstrate how an inclusive exon microarray can be used to discover novel alternative splicing events. As examples, 17 new tissue-specific exons from 11 genes were validated by RT-PCR and sequencing.
Conclusion: In conjunction with a conceptually simple algorithm, comprehensive exon microarrays can detect tissue-specific alternative splicing events. Our data suggest significant expression outside of known exons and well annotated genes and a high frequency of alternative splicing events. In addition, we identified and validated a number of novel exons with tissue-specific splicing patterns. The tissue map data will likely serve as a valuable source of information on the regulation of alternative splicing.
Figures




References
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

