Complex chromosome 17p rearrangements associated with low-copy repeats in two patients with congenital anomalies
- PMID: 17457615
- PMCID: PMC1914245
- DOI: 10.1007/s00439-007-0359-6
Complex chromosome 17p rearrangements associated with low-copy repeats in two patients with congenital anomalies
Abstract
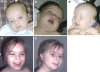
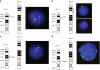
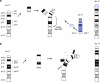
Recent molecular cytogenetic data have shown that the constitution of complex chromosome rearrangements (CCRs) may be more complicated than previously thought. The complicated nature of these rearrangements challenges the accurate delineation of the chromosomal breakpoints and mechanisms involved. Here, we report a molecular cytogenetic analysis of two patients with congenital anomalies and unbalanced de novo CCRs involving chromosome 17p using high-resolution array-based comparative genomic hybridization (array CGH) and fluorescent in situ hybridization (FISH). In the first patient, a 4-month-old boy with developmental delay, hypotonia, growth retardation, coronal synostosis, mild hypertelorism, and bilateral club feet, we found a duplication of the Charcot-Marie-Tooth disease type 1A and Smith-Magenis syndrome (SMS) chromosome regions, inverted insertion of the Miller-Dieker lissencephaly syndrome region into the SMS region, and two microdeletions including a terminal deletion of 17p. The latter, together with a duplication of 21q22.3-qter detected by array CGH, are likely the unbalanced product of a translocation t(17;21)(p13.3;q22.3). In the second patient, an 8-year-old girl with mental retardation, short stature, microcephaly and mild dysmorphic features, we identified four submicroscopic interspersed 17p duplications. All 17 breakpoints were examined in detail by FISH analysis. We found that four of the breakpoints mapped within known low-copy repeats (LCRs), including LCR17pA, middle SMS-REP/LCR17pB block, and LCR17pC. Our findings suggest that the LCR burden in proximal 17p may have stimulated the formation of these CCRs and, thus, that genome architectural features such as LCRs may have been instrumental in the generation of these CCRs.
Figures



References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1007/s00439-003-1079-1', 'is_inner': False, 'url': 'https://doi.org/10.1007/s00439-003-1079-1'}, {'type': 'PubMed', 'value': '14767757', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/14767757/'}]}
- Astbury C, Christ LA, Aughton DJ, Cassidy SB, Fujimoto A, Pletcher BA, Schafer IA, Schwartz S (2004) Delineation of complex chromosomal rearrangements: evidence for increased complexity. Hum Genet 114:448–457 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1038/ng0306-269', 'is_inner': False, 'url': 'https://doi.org/10.1038/ng0306-269'}, {'type': 'PubMed', 'value': '16940994', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/16940994/'}]}
- Axton M (2006) Structural variants deconstruct the genome. Nat Genet 38:959 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1101/gr.GR-1871R', 'is_inner': False, 'url': 'https://doi.org/10.1101/gr.gr-1871r'}, {'type': 'PMC', 'value': 'PMC311093', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC311093/'}, {'type': 'PubMed', 'value': '11381028', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/11381028/'}]}
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11:1005–1017 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1126/science.1072047', 'is_inner': False, 'url': 'https://doi.org/10.1126/science.1072047'}, {'type': 'PubMed', 'value': '12169732', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12169732/'}]}
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297:1003–1007 - PubMed
-
- Bartels I, Starke H, Argyriou L, Sauter SM, Zoll B, Liehr T (2006) An exceptional complex chromosomal rearrangement (CCR) with eight breakpoints involving four chromosomes (1;3;9;14) in an azoospermic male with normal phenotype. Eur J Med Genet doi:10.1016/j.ejmg.2006.10.007 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

