Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation
- PMID: 17459921
- PMCID: PMC1933317
- DOI: 10.1128/JVI.00029-07
Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation
Abstract
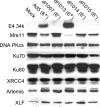
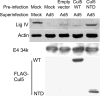
Cells infected by adenovirus E4 mutants accumulate end-to-end concatemers of the viral genome that are assembled from unit-length viral DNAs by nonhomologous end joining (NHEJ). Genome concatenation can be prevented by expression either of E4 11k (product of E4orf3) or of the complex of E4 34k (product of E4orf6) and E1b 55k. Both E4 11k and the E4 34k/E1b 55k complex prevent concatenation at least in part by inactivation of the host protein Mre11: E4 11k sequesters Mre11 in aggresomes, while the E4 34k/E1b 55k complex participates in a virus-specific E3 ubiquitin ligase that mediates ubiquitination and proteasomal degradation. The E4 34k/E1b 55k complex, but not E4 11k, also inhibits NHEJ activity on internal breaks in the viral genome and on V(D)J recombination substrate plasmids, suggesting that it may interfere with NHEJ independently of its effect on Mre11. We show here that DNA ligase IV, which performs the joining step of NHEJ, is degraded as a consequence of adenovirus infection. Degradation is dependent upon E4 34k and E1b 55k, functional proteasomes, and the activity of cellular cullin 5, a component of the adenoviral ubiquitin ligase. DNA ligase IV also interacts physically with E1b 55k. The data demonstrate that DNA ligase IV, like Mre11, is a substrate for the adenovirus-specific E3 ubiquitin ligase; identify an additional viral approach to prevention of genome concatenation; and provide a mechanism for the general inhibition of NHEJ by adenoviruses.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

