Atypical bZIP domain of viral transcription factor contributes to stability of dimer formation and transcriptional function
- PMID: 17459922
- PMCID: PMC1933325
- DOI: 10.1128/JVI.00215-07
Atypical bZIP domain of viral transcription factor contributes to stability of dimer formation and transcriptional function
Abstract
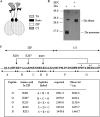
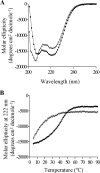
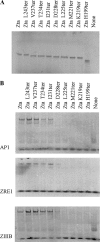
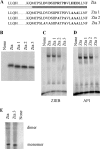
The Epstein-Barr virus transcription factor Zta (encoded by BZLF1) is a bZIP protein containing an alpha-helical coiled-coil homodimerization motif (zipper). The Zta zipper forms less-stable dimers than other bZIP proteins, and an adjacent region (CT) interacts with the zipper to form a novel structure that is proposed to strengthen the dimer. Here we question the role of the CT region for Zta function. Cross-linking experiments demonstrate that the entire CT region lies adjacent to the zipper. Detailed analyses of Zta truncation mutations identify an involvement of the proximal CT region (221 to 230) in dimer formation with a further contribution from the distal region (236 to 243). Biophysical analyses reveal that residues 221 to 230 enhance the stability of the coiled coil. The ability of the Zta truncation mutants to interact with three Zta-binding sites also requires the proximal CT region. Fine mapping of DNA-binding requirements highlighted the contribution of these amino acids for Zta function. Thus, the proximal part of the CT region is required to aid the dimerization of Zta and thereby its DNA-binding ability. In contrast, although the distal part of the CT region aids dimerization, it promotes only a modest increase in DNA binding. To probe this further, we defined the contribution from the CT region for Zta to transactivate a promoter embedded within the viral genome. From this we conclude that the proximal part of the CT region is absolutely required, whereas the distal part is dispensable.
Figures


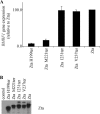
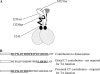
References
-
- Al Mehairi, S., E. Cerasoli, and A. J. Sinclair. 2005. Investigation of the multimerization region of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) protein K-bZIP: the proposed leucine zipper region encodes a multimerization domain with an unusual structure. J. Virol. 79:7905-7910. - PMC - PubMed
-
- Bhende, P. M., W. T. Seaman, H. J. Delecluse, and S. C. Kenney. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 36:1099-1104. - PubMed
-
- Cayrol, C., and E. Flemington. 1996. G(0)/G(1), growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J. Biol. Chem. 271:31799-31802. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

