Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125
- PMID: 17460044
- PMCID: PMC1863485
- DOI: 10.1073/pnas.0611551104
Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125
Abstract
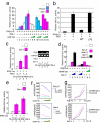
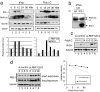
Retinoic acid-inducible gene I (RIG-I) plays a pivotal role in the regulation of cytokine production induced by pathogens. The RIG-I also augments the production of IFN and other cytokines via an amplification circuit. Because the production of cytokines is closely controlled, up- and down-regulation of RIG-I signaling also needs strict regulation. The mechanism of this regulation, however, remains elusive. Here, we found that RIG-I undergoes proteasomal degradation after conjugation to ubiquitin by RNF125. Further, RNF125 conjugates ubiquitin to MDA5, a family protein of RIG-I as well as IPS-1, which is also a downstream protein of RIG-I signaling that results in suppressing the functions of these proteins. Because RNF125 is enhanced by IFN, these functions constitute a negative regulatory loop circuit for IFN production.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Iwasaki A, Medzhitov R. Nat Immunol. 2004;5:987–995. - PubMed
-
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Annu Rev Immunol. 2005;23:307–336. - PubMed
-
- Takeda K, Akira S. Int Immunol. 2005;17:1–14. - PubMed
-
- Medzhitov R, Janeway CA., Jr Cell. 1997;91:295–298. - PubMed
-
- Janeway CA, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

