Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression
- PMID: 17461777
- PMCID: PMC2267305
- DOI: 10.1042/BJ20061812
Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression
Abstract
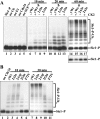
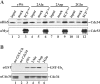
The ubiquitin-conjugating enzyme Cdc34 (cell division cycle 34) plays an essential role in promoting the G1-S-phase transition of the eukaryotic cell cycle and is phosphorylated in vivo. In the present study, we investigated if phosphorylation regulates Cdc34 function. We mapped the in vivo phosphorylation sites on budding yeast Cdc34 (yCdc34; Ser207 and Ser216) and human Cdc34 (hCdc34 Ser203, Ser222 and Ser231) to serine residues in the acidic tail domain, a region that is critical for Cdc34's cell cycle function. CK2 (protein kinase CK2) phosphorylates both yCdc34 and hCdc34 on these sites in vitro. CK2-mediated phosphorylation increased yCdc34 ubiquitination activity towards the yeast Saccharomyces cerevisiae Sic1 in vitro, when assayed in the presence of its cognate SCFCdc4 E3 ligase [where SCF is Skp1 (S-phase kinase-associated protein 1)/cullin/F-box]. Similarly, mutation of the yCdc34 phosphorylation sites to alanine, aspartate or glutamate residues altered Cdc34-SCFCdc4-mediated Sic1 ubiquitination activity. Similar results were obtained when yCdc34's ubiquitination activity was assayed in the absence of SCFCdc4, indicating that phosphorylation regulates the intrinsic catalytic activity of Cdc34. To evaluate the in vivo consequences of altered Cdc34 activity, wild-type yCdc34 and the phosphosite mutants were introduced into an S. cerevisiae cdc34 deletion strain and, following synchronization in G1-phase, progression through the cell cycle was monitored. Consistent with the increased ubiquitination activity in vitro, cells expressing the phosphosite mutants with higher catalytic activity exhibited accelerated cell cycle progression and Sic1 degradation. These studies demonstrate that CK2-mediated phosphorylation of Cdc34 on the acidic tail domain stimulates Cdc34-SCFCdc4 ubiquitination activity and cell cycle progression.
Figures


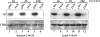

Similar articles
-
The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4.J Biol Chem. 2009 Dec 25;284(52):36012-36023. doi: 10.1074/jbc.M109.058529. Epub 2009 Oct 29. J Biol Chem. 2009. PMID: 19875449 Free PMC article.
-
SCF E3-mediated autoubiquitination negatively regulates activity of Cdc34 E2 but plays a nonessential role in the catalytic cycle in vitro and in vivo.Mol Cell Biol. 2007 Aug;27(16):5860-70. doi: 10.1128/MCB.01555-06. Epub 2007 Jun 11. Mol Cell Biol. 2007. PMID: 17562869 Free PMC article.
-
Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34.Mol Cell Biol. 2010 May;30(10):2316-29. doi: 10.1128/MCB.01094-09. Epub 2010 Mar 1. Mol Cell Biol. 2010. PMID: 20194622 Free PMC article.
-
Ubiquitin ligases and cell cycle control.Annu Rev Biochem. 2013;82:387-414. doi: 10.1146/annurev-biochem-060410-105307. Epub 2013 Mar 13. Annu Rev Biochem. 2013. PMID: 23495935 Review.
-
Structure, function and mechanism of the anaphase promoting complex (APC/C).Q Rev Biophys. 2011 May;44(2):153-90. doi: 10.1017/S0033583510000259. Epub 2010 Nov 22. Q Rev Biophys. 2011. PMID: 21092369 Review.
Cited by
-
Mechanisms of generating polyubiquitin chains of different topology.Cells. 2014 Jul 1;3(3):674-89. doi: 10.3390/cells3030674. Cells. 2014. PMID: 24987835 Free PMC article.
-
The fatty acid synthase inhibitor triclosan: repurposing an anti-microbial agent for targeting prostate cancer.Oncotarget. 2014 Oct 15;5(19):9362-81. doi: 10.18632/oncotarget.2433. Oncotarget. 2014. PMID: 25313139 Free PMC article.
-
Characterization of the complex involved in regulating V-ATPase activity of the vacuolar and endosomal membrane.J Bioenerg Biomembr. 2017 Oct;49(5):347-355. doi: 10.1007/s10863-017-9712-1. Epub 2017 Jun 22. J Bioenerg Biomembr. 2017. PMID: 28643238
-
Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine.Cell Div. 2010 Aug 13;5:19. doi: 10.1186/1747-1028-5-19. Cell Div. 2010. PMID: 20704751 Free PMC article.
-
Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination.FEBS J. 2009 Oct;276(19):5381-9. doi: 10.1111/j.1742-4658.2009.07249.x. Epub 2009 Aug 27. FEBS J. 2009. PMID: 19712108 Free PMC article. Review.
References
-
- Pickart C., Eddins M. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. - PubMed
-
- Sun L., Chen Z. J. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 2004;16:119–126. - PubMed
-
- Pickart C. M., Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. - PubMed
-
- Petroski M., Deshaies R. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:8–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

