An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin
- PMID: 17462020
- PMCID: PMC1891008
- DOI: 10.1111/j.1365-2958.2007.05688.x
An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin
Abstract
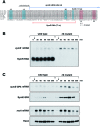
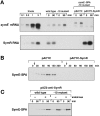
Only few small, regulatory RNAs encoded opposite another gene have been identified in bacteria. Here, we report the characterization of a locus where a small RNA (SymR) is encoded in cis to an SOS-induced gene whose product shows homology to the antitoxin MazE (SymE). Synthesis of the SymE protein is tightly repressed at multiple levels by the LexA repressor, the SymR RNA and the Lon protease. SymE co-purifies with ribosomes and overproduction of the protein leads to cell growth inhibition, decreased protein synthesis and increased RNA degradation. These properties are shared with several RNA endonuclease toxins of the toxin-antitoxin modules, and we show that the SymE protein represents evolution of a toxin from the AbrB fold, whose representatives are typically antitoxins. We suggest that SymE promotion of RNA cleavage may be important for the recycling of RNAs damaged under SOS-inducing conditions.
Figures





Similar articles
-
Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR.RNA Biol. 2012 Dec;9(12):1520-7. doi: 10.4161/rna.22757. Epub 2012 Nov 6. RNA Biol. 2012. PMID: 23131729 Review.
-
RNA antitoxins.Curr Opin Microbiol. 2007 Apr;10(2):117-24. doi: 10.1016/j.mib.2007.03.003. Epub 2007 Mar 21. Curr Opin Microbiol. 2007. PMID: 17376733 Review.
-
Toxin-antitoxin modules as bacterial metabolic stress managers.Trends Biochem Sci. 2005 Dec;30(12):672-9. doi: 10.1016/j.tibs.2005.10.004. Epub 2005 Oct 28. Trends Biochem Sci. 2005. PMID: 16257530 Review.
-
Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes.J Mol Biol. 1999 Jan 29;285(4):1401-15. doi: 10.1006/jmbi.1998.2416. J Mol Biol. 1999. PMID: 9917385
-
An antisense RNA inhibits translation by competing with standby ribosomes.Mol Cell. 2007 May 11;26(3):381-92. doi: 10.1016/j.molcel.2007.04.003. Mol Cell. 2007. PMID: 17499044
Cited by
-
Regulation of polyphosphate kinase production by antisense RNA in Pseudomonas fluorescens Pf0-1.Appl Environ Microbiol. 2012 Jun;78(12):4533-7. doi: 10.1128/AEM.07836-11. Epub 2012 Apr 6. Appl Environ Microbiol. 2012. PMID: 22492458 Free PMC article.
-
Functional genomics reveals the toxin-antitoxin repertoire and AbiE activity in Serratia.Microb Genom. 2020 Nov;6(11):mgen000458. doi: 10.1099/mgen.0.000458. Microb Genom. 2020. PMID: 33074086 Free PMC article.
-
Topologies of synthetic gene circuit for optimal fold change activation.Nucleic Acids Res. 2021 May 21;49(9):5393-5406. doi: 10.1093/nar/gkab253. Nucleic Acids Res. 2021. PMID: 34009384 Free PMC article.
-
The small RNA SraG participates in PNPase homeostasis.RNA. 2016 Oct;22(10):1560-73. doi: 10.1261/rna.055236.115. Epub 2016 Aug 5. RNA. 2016. PMID: 27495318 Free PMC article.
-
Chromosomally Encoded hok-sok Toxin-Antitoxin System in the Fire Blight Pathogen Erwinia amylovora: Identification and Functional Characterization.Appl Environ Microbiol. 2019 Jul 18;85(15):e00724-19. doi: 10.1128/AEM.00724-19. Print 2019 Aug 1. Appl Environ Microbiol. 2019. PMID: 31101613 Free PMC article.
References
-
- Anantharaman V, Aravind L. Novel predicted RNAses with a PIN Domain-like Fold. RNA Biol. 2006;3:18–27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

