Guanine nucleotide depletion inhibits pre-ribosomal RNA synthesis and causes nucleolar disruption
- PMID: 17462731
- PMCID: PMC4552191
- DOI: 10.1016/j.leukres.2007.03.025
Guanine nucleotide depletion inhibits pre-ribosomal RNA synthesis and causes nucleolar disruption
Abstract
Inosine monophosphate dehydrogenase (IMPDH) is a pivotal enzyme in the de novo pathway of guanine nucleotide biosynthesis. Inhibitors of this enzyme decrease intracellular guanine nucleotide levels by 50-80% and have potential as anti-neoplastic agents. Both mycophenolic acid (MPA) and AVN-944 are highly specific inhibitors of IMPDH that cause cell cycle arrest or apoptosis in lymphocytes and leukemic cell lines. We have examined the mechanisms by which these two agents cause cytotoxicity. Both MPA and AVN-944 inhibit the growth of K562 cells, and induce apoptosis in Raji B and CCRF-CEM T cells. Both compounds strikingly inhibit RNA synthesis within 2 h of exposure. Depletion of guanine nucleotides by MPA and AVN-944 also causes an early and near-complete reduction in levels of the 45S precursor rRNA synthesis and the concomitant translocation of nucleolar proteins including nucleolin, nucleophosmin, and nucleostemin from the nucleolus to the nucleoplasm. This efflux correlates temporally with the sustained induction of p53 in cell lines with wild-type p53. We conclude that inhibition of IMPDH causes a primary reduction in rRNA synthesis and secondary nucleolar disruption and efflux of nucleolar proteins that most likely mediate cell cycle arrest or apoptosis. The ability of AVN-944 to induce apoptosis in a number of leukemic cell lines supports its potential utility in the treatment of hematologic malignancies.
Figures








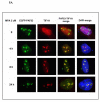

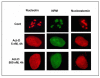
References
-
- Zimmermann AG, Gu JJ, Laliberte J, et al. Inosine-5′-monophosphate dehydrogenase: regulation of expression and role in cellular proliferation and T lymphocyte activation. Prog Nucleic Acid Res Mol Biol. 1998;61:181–209. - PubMed
-
- Allison AC, Eugui EM. Mechanisms of action of mycophenolate mofetil in preventing acute and chronic allograft rejection. Transplantation. 2005;80:S181–190. - PubMed
-
- Laliberte J, Yee A, Xiong Y, et al. Effects of guanine nucleotide depletion on cell cycle progression in human T lymphocytes. Blood. 1998;91:2896–2904. - PubMed
-
- Heinschink A, Raab M, Daxecker H, et al. In vitro effects of mycophenolic acid on cell cycle and activation of human lymphocytes. Clin Chim Acta. 2000;300:23–28. - PubMed
-
- Yalowitz JA, Pankiewicz K, Patterson SE, et al. Cytotoxicity and cellular differentiation activity of methylenebis(phosphonate) analogs of tiazofurin and mycophenolic acid adenine dinucleotide in human cancer cell lines. Cancer Lett. 2002;181:31–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

