Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation
- PMID: 17463081
- PMCID: PMC1857225
- DOI: 10.1073/pnas.0702609104
Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation
Abstract
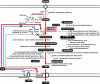
The L-valine production strain of Escherichia coli was constructed by rational metabolic engineering and stepwise improvement based on transcriptome analysis and gene knockout simulation of the in silico genome-scale metabolic network. Feedback inhibition of acetohydroxy acid synthase isoenzyme III by L-valine was removed by site-directed mutagenesis, and the native promoter containing the transcriptional attenuator leader regions of the ilvGMEDA and ilvBN operon was replaced with the tac promoter. The ilvA, leuA, and panB genes were deleted to make more precursors available for L-valine biosynthesis. This engineered Val strain harboring a plasmid overexpressing the ilvBN genes produced 1.31 g/liter L-valine. Comparative transcriptome profiling was performed during batch fermentation of the engineered and control strains. Among the down-regulated genes, the lrp and ygaZH genes, which encode a global regulator Lrp and L-valine exporter, respectively, were overexpressed. Amplification of the lrp, ygaZH, and lrp-ygaZH genes led to the enhanced production of L-valine by 21.6%, 47.1%, and 113%, respectively. Further improvement was achieved by using in silico gene knockout simulation, which identified the aceF, mdh, and pfkA genes as knockout targets. The VAMF strain (Val DeltaaceF Deltamdh DeltapfkA) overexpressing the ilvBN, ilvCED, ygaZH, and lrp genes was able to produce 7.55 g/liter L-valine from 20 g/liter glucose in batch culture, resulting in a high yield of 0.378 g of L-valine per gram of glucose. These results suggest that an industrially competitive strain can be efficiently developed by metabolic engineering based on combined rational modification, transcriptome profiling, and systems-level in silico analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lee SY, Lee DY, Kim TY. Trends Biotechnol. 2005;23:349–358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

