Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain
- PMID: 17464284
- PMCID: PMC1868911
- DOI: 10.1038/sj.emboj.7601700
Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain
Abstract
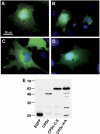
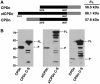
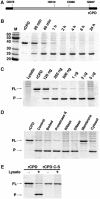
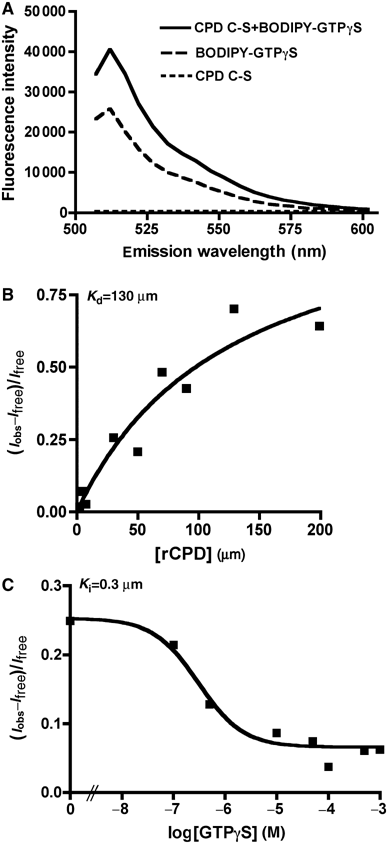
Vibrio cholerae RTX is a large multifunctional bacterial toxin that causes actin crosslinking. Due to its size, it was predicted to undergo proteolytic cleavage during translocation into host cells to deliver activity domains to the cytosol. In this study, we identified a domain within the RTX toxin that is conserved in large clostridial glucosylating toxins TcdB, TcdA, TcnA, and TcsL; putative toxins from V. vulnificus, Yersinia sp., Photorhabdus sp., and Xenorhabdus sp.; and a filamentous/hemagglutinin-like protein FhaL from Bordetella sp. In vivo transfection studies and in vitro characterization of purified recombinant protein revealed that this domain from the V. cholerae RTX toxin is an autoprocessing cysteine protease whose activity is stimulated by the intracellular environment. A cysteine point mutation within the RTX holotoxin attenuated actin crosslinking activity suggesting that processing of the toxin is an important step in toxin translocation. Overall, we have uncovered a new mechanism by which large bacterial toxins and proteins deliver catalytic activities to the eukaryotic cell cytosol by autoprocessing after translocation.
Figures



References
-
- Anderson BM, Vasini EC (1970) Nonpolar effects in reactions of the sulfhydryl group of papain. Biochemistry 9: 3348–3352 - PubMed
-
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in bioploymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp 28–36. Menlo Park, CA: AAAI Press - PubMed
-
- Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54 - PubMed
-
- Barroso LA, Moncrief JS, Lyerly DM, Wilkins TD (1994) Mutagenesis of the Clostridium difficile toxin B gene and effect on cytotoxic activity. Microb Pathog 16: 297–303 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

