The lin-35/Rb and RNAi pathways cooperate to regulate a key cell cycle transition in C. elegans
- PMID: 17466069
- PMCID: PMC1877806
- DOI: 10.1186/1471-213X-7-38
The lin-35/Rb and RNAi pathways cooperate to regulate a key cell cycle transition in C. elegans
Abstract
Background: The Retinoblastoma gene product (Rb) has been shown to regulate the transcription of key genes involved in cell growth and proliferation. Consistent with this, mutations in Rb are associated with numerous types of cancer making it a critical tumour suppressor gene. Its function is conferred through a large multiprotein complex that exhibits a dual function in both activation and repression of gene targets. In C. elegans, the Rb orthologue lin-35 functions redundantly with other transcriptional regulators to appropriately specify both vulval and pharyngeal cell fates.
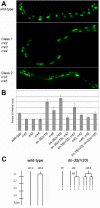
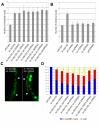
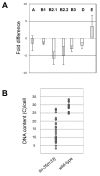
Results: In C. elegans the intestinal cells must alter their cell cycle from the mitotic cell divisions typical of embryogenesis to karyokinesis and then endoreplication, which facilitates growth during larval development. While screening for genes that affect the ability of the intestinal cells to appropriately make this cell cycle transition during post-embryonic development, we isolated mutants that either compromise this switch and remain mononucleate, or cause these cells to undergo multiple rounds of nuclear division. Among these mutants we identified a novel allele of lin-35/Rb, while we also found that the components of the synMuv B complex, which are involved in vulval specification, are also required to properly regulate the developmentally-controlled cell cycle transition typical of these intestinal cells during larval development. More importantly, our work uncovered a role for certain members of the pathways involved in RNAi in mediating the efficient transition between these cell cycle programs, suggesting that lin-35/Rb cooperates with these RNAi components. Furthermore, our findings suggest that met-2, a methyltransferase as well as hpl-1 and hpl-2, two C. elegans homologues of the heterochromatin protein HP1 are also required for this transition.
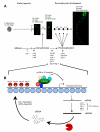
Conclusion: Our findings are consistent with lin-35/Rb, synMuv and RNAi components cooperating, probably through their additive effects on chromatin modification, to appropriately modulate the expression of genes that are required to switch from the karyokinesis cell cycle to endoreplication; a highly specified growth pathway in the intestinal epithelium. The lin-35/Rb repressor complex may be required to initiate this process, while components of the RNAi machinery positively reinforce this repression.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

