Understanding the isomerization of the HIV-1 dimerization initiation domain by the nucleocapsid protein
- PMID: 17466332
- PMCID: PMC2475603
- DOI: 10.1016/j.jmb.2007.03.065
Understanding the isomerization of the HIV-1 dimerization initiation domain by the nucleocapsid protein
Abstract
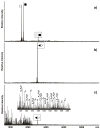
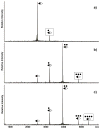
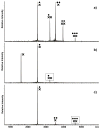
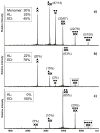
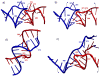
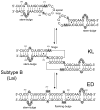
The specific binding of HIV-1 nucleocapsid (NC) to the hinge region of the kissing-loop (KL) dimer formed by stemloop 1 (SL1) can have significant consequences on its ability to isomerize into the corresponding extended duplex (ED) form. The binding determinants and the effects on the isomerization process were investigated in vitro by a concerted strategy involving ad hoc RNA mutants and electrospray ionization-Fourier transform ion cyclotron resonance (ESI-FTICR) mass spectrometry, which enabled us to characterize the stoichiometry and conformational state of all possible protein-RNA and RNA-RNA assemblies present simultaneously in solution. For the first time, NC-hinge interactions were observed in constructs including at least one unpaired guanine at the 5'-end of the loop-loop duplex, whereas no interactions were detected when the unpaired guanine was placed at its 3'-end. This binding mode is supported by the presence of a grip-like motif described by recent crystal structures, which is formed by the 5'-purines of both hairpins held together by mutual stacking interactions. Using tandem mass spectrometry, hinge interactions were clearly shown to reduce the efficiency of KL/ED isomerization without inducing its complete block. This outcome is consistent with the partial stabilization of the extra-helical grip by the bound protein, which can hamper the purine components from parting ways and initiate the strand exchange process. These findings confirm that the broad binding and chaperone activities of NC induce unique effects that are clearly dependent on the structural context of the cognate nucleic acid substrate. For this reason, the presence of multiple binding sites on the different forms assumed by SL1 can produce seemingly contrasting effects that contribute to a fine modulation of the two-step process of RNA dimerization and isomerization.
Figures


Similar articles
-
Noncovalent probes for the investigation of structure and dynamics of protein-nucleic acid assemblies: the case of NC-mediated dimerization of genomic RNA in HIV-1.Biopolymers. 2009 Apr;91(4):283-96. doi: 10.1002/bip.21107. Biopolymers. 2009. PMID: 18946871 Free PMC article.
-
Dissecting the protein-RNA and RNA-RNA interactions in the nucleocapsid-mediated dimerization and isomerization of HIV-1 stemloop 1.J Mol Biol. 2007 Jan 12;365(2):396-410. doi: 10.1016/j.jmb.2006.09.081. Epub 2006 Oct 3. J Mol Biol. 2007. PMID: 17070549 Free PMC article.
-
Functional investigations of retroviral proteinribonucleic acid complexes by nanospray Fourier transform ion cyclotron resonance mass spectrometry.Eur J Mass Spectrom (Chichester). 2007;13(1):29-33. doi: 10.1255/ejms.839. Eur J Mass Spectrom (Chichester). 2007. PMID: 17878535
-
Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1.Biochemistry. 2002 Dec 17;41(50):14762-70. doi: 10.1021/bi0267240. Biochemistry. 2002. PMID: 12475224
-
Direct probing of RNA structures and RNA-protein interactions in the HIV-1 packaging signal by chemical modification and electrospray ionization fourier transform mass spectrometry.J Mol Biol. 2003 Jul 4;330(2):211-23. doi: 10.1016/s0022-2836(03)00589-8. J Mol Biol. 2003. PMID: 12823962
Cited by
-
SL1 revisited: functional analysis of the structure and conformation of HIV-1 genome RNA.Retrovirology. 2016 Nov 11;13(1):79. doi: 10.1186/s12977-016-0310-9. Retrovirology. 2016. PMID: 27835956 Free PMC article.
-
Direct and Topoisomerase II Mediated DNA Damage by Bis-3-chloropiperidines: The Importance of Being an Earnest G.ChemMedChem. 2017 Sep 7;12(17):1471-1479. doi: 10.1002/cmdc.201700368. Epub 2017 Aug 10. ChemMedChem. 2017. PMID: 28724198 Free PMC article.
-
A role for the MS analysis of nucleic acids in the post-genomics age.J Am Soc Mass Spectrom. 2010 Jan;21(1):1-13. doi: 10.1016/j.jasms.2009.09.006. Epub 2009 Sep 17. J Am Soc Mass Spectrom. 2010. PMID: 19897384
-
HIV-1 DIS stem loop forms an obligatory bent kissing intermediate in the dimerization pathway.Nucleic Acids Res. 2014 Jun;42(11):7281-9. doi: 10.1093/nar/gku332. Epub 2014 May 9. Nucleic Acids Res. 2014. PMID: 24813449 Free PMC article.
-
Visualizing transient low-populated structures of RNA.Nature. 2012 Nov 29;491(7426):724-8. doi: 10.1038/nature11498. Epub 2012 Oct 7. Nature. 2012. PMID: 23041928 Free PMC article.
References
-
- Laughrea M, Jetté L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. - PubMed
-
- Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–653. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

