L-sorbose reductase and its transcriptional regulator involved in L-sorbose utilization of Gluconobacter frateurii
- PMID: 17468249
- PMCID: PMC1913458
- DOI: 10.1128/JB.01895-06
L-sorbose reductase and its transcriptional regulator involved in L-sorbose utilization of Gluconobacter frateurii
Abstract
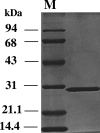
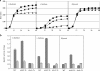
Upstream of the gene for flavin adenine dinucleotide (FAD)-dependent D-sorbitol dehydrogenase (SLDH), sldSLC, a putative transcriptional regulator was found in Gluconobacter frateurii THD32 (NBRC 101656). In this study, the whole sboR gene and the adjacent gene, sboA, were cloned and analyzed. sboR mutation did not affect FAD-SLDH activity in the membrane fractions. The SboA enzyme expressed and purified from an Escherichia coli transformant showed NADPH-dependent L-sorbose reductase (NADPH-SR) activity, and the enzyme was different from the NADPH-SR previously reported for Gluconobacter suboxydans IFO 3291 in molecular size and amino acid sequence. A mutant defective in sboA showed significantly reduced growth on L-sorbose, indicating that the SboA enzyme is required for efficient growth on L-sorbose. The sboR mutant grew on L-sorbose even better than the wild-type strain did, and higher NADPH-SR activity was detected in cytoplasm fractions. Reverse transcription-PCR experiments indicated that sboRA comprises an operon. These data suggest that sboR is involved in the repression of sboA, but not in the induction of sldSLC, on D-sorbitol and that another activator is required for the induction of these genes by D-sorbitol or L-sorbose.
Figures



Similar articles
-
Molecular properties of membrane-bound FAD-containing D-sorbitol dehydrogenase from thermotolerant Gluconobacter frateurii isolated from Thailand.Biosci Biotechnol Biochem. 2005 Jun;69(6):1120-9. doi: 10.1271/bbb.69.1120. Biosci Biotechnol Biochem. 2005. PMID: 15973043
-
High-temperature sorbose fermentation with thermotolerant Gluconobacter frateurii CHM43 and its mutant strain adapted to higher temperature.Appl Microbiol Biotechnol. 2012 Sep;95(6):1531-40. doi: 10.1007/s00253-012-4005-4. Epub 2012 Mar 22. Appl Microbiol Biotechnol. 2012. PMID: 22434571
-
Characterization of genes involved in D-sorbitol oxidation in thermotolerant Gluconobacter frateurii.Biosci Biotechnol Biochem. 2012;76(8):1497-505. doi: 10.1271/bbb.120227. Epub 2012 Aug 7. Biosci Biotechnol Biochem. 2012. PMID: 22878210
-
New developments in oxidative fermentation.Appl Microbiol Biotechnol. 2003 Feb;60(6):643-53. doi: 10.1007/s00253-002-1155-9. Epub 2002 Dec 18. Appl Microbiol Biotechnol. 2003. PMID: 12664142 Review.
-
New quinoproteins in oxidative fermentation.Biochim Biophys Acta. 2003 Apr 11;1647(1-2):10-7. doi: 10.1016/s1570-9639(03)00040-2. Biochim Biophys Acta. 2003. PMID: 12686101 Review.
Cited by
-
Characterization of a novel D-sorbitol dehydrogenase from Faunimonas pinastri A52C2.Appl Microbiol Biotechnol. 2025 Jan 27;109(1):25. doi: 10.1007/s00253-024-13381-2. Appl Microbiol Biotechnol. 2025. PMID: 39869196 Free PMC article.
-
Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation.Microb Cell Fact. 2016 Jan 25;15:21. doi: 10.1186/s12934-016-0418-6. Microb Cell Fact. 2016. PMID: 26809519 Free PMC article.
-
Purification, crystallization and preliminary X-ray analysis of L-sorbose reductase from Gluconobacter frateurii complexed with L-sorbose or NADPH.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Jun 1;65(Pt 6):562-4. doi: 10.1107/S1744309109014687. Epub 2009 May 22. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19478431 Free PMC article.
-
On the way toward regulatable expression systems in acetic acid bacteria: target gene expression and use cases.Appl Microbiol Biotechnol. 2021 May;105(9):3423-3456. doi: 10.1007/s00253-021-11269-z. Epub 2021 Apr 15. Appl Microbiol Biotechnol. 2021. PMID: 33856535 Free PMC article. Review.
References
-
- Adachi, O., H. Toyama, G. Theeragool, N. Lotong, and K. Matsushita. 1999. Crystallization and properties of NAD-dependent d-sorbitol dehydrogenase from Gluconobacter suboxydans IFO 3257. Biosci. Biotechnol. Biochem. 63:1589-1595. - PubMed
-
- Adachi, O., H. Toyama, G. Theeragool, N. Lotong, and K. Matsushita. 1999. Crystallization and properties of NADPH-dependent l-sorbose reductase from Gluconobacter melanogenus IFO 3294. Biosci. Biotechnol. Biochem. 63:2137-2143. - PubMed
-
- Adachi, O., D. Moonmungmee, E. Shinagawa, H. Toyama, M. Yamada, and K. Matsushita. 2003. New quinoprotein in oxidative fermentation. Biochim. Biophys. Acta 1647:10-17. - PubMed
-
- Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

