Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development
- PMID: 17468259
- PMCID: PMC1913762
- DOI: 10.1105/tpc.107.051375
Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development
Abstract
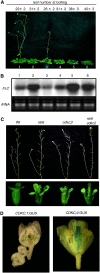
The C-terminal domain (CTD) of RNA polymerase II is phosphorylated during the transcription cycle by three cyclin-dependent kinases (CDKs): CDK7, CDK8, and CDK9. CDK9 and its interacting cyclin T partners belong to the positive transcription elongation factor b (P-TEFb) complexes, which phosphorylate the CTD to promote transcription elongation. We report that Arabidopsis thaliana CDK9-like proteins, CDKC;1 and CDKC;2, and their interacting cyclin T partners, CYCT1;4 and CYCT1;5, play important roles in infection with Cauliflower mosaic virus (CaMV). cdkc;2 and cyct1;5 knockout mutants are highly resistant and cdkc;2 cyct1;5 double mutants are extremely resistant to CaMV. The mutants respond normally to other types of plant viruses that do not replicate by reverse transcription. Expression of a reporter gene driven by the CaMV 35S promoter is markedly reduced in the cdkc;2 and cyct1;5 mutants, indicating that the kinase complexes are important for transcription from the viral promoter. Loss of function of CDKC;1/CDKC;2 or CYCT1;4/CYCT1;5 results in complete resistance to CaMV as well as altered leaf and flower growth, trichome development, and delayed flowering. These results establish Arabidopsis CDKC kinase complexes as important host targets of CaMV for transcriptional activation of viral genes and critical regulators of plant growth and development.
Figures

References
-
- Adamson, T.E., Shutt, D.C., and Price, D.H. (2005). Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 280 32262–32271. - PubMed
-
- Akoulitchev, S., Chuikov, S., and Reinberg, D. (2000). TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407 102–106. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

