A rapid and efficient method for cloning genes of type II restriction-modification systems by use of a killer plasmid
- PMID: 17468281
- PMCID: PMC1932789
- DOI: 10.1128/AEM.00119-07
A rapid and efficient method for cloning genes of type II restriction-modification systems by use of a killer plasmid
Abstract
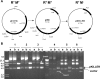
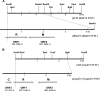
We present a method for cloning restriction-modification (R-M) systems that is based on the use of a lethal plasmid (pKILLER). The plasmid carries a functional gene for a restriction endonuclease having the same DNA specificity as the R-M system of interest. The first step is the standard preparation of a representative, plasmid-borne genomic library. Then this library is transformed with the killer plasmid. The only surviving bacteria are those which carry the gene specifying a protective DNA methyltransferase. Conceptually, this in vivo selection approach resembles earlier methods in which a plasmid library was selected in vitro by digestion with a suitable restriction endonuclease, but it is much more efficient than those methods. The new method was successfully used to clone two R-M systems, BstZ1II from Bacillus stearothermophilus 14P and Csp231I from Citrobacter sp. strain RFL231, both isospecific to the prototype HindIII R-M system.
Figures

Similar articles
-
Characterization of pEC156, a ColE1-type plasmid from Escherichia coli E1585-68 that carries genes of the EcoVIII restriction-modification system.Plasmid. 2001 Sep;46(2):128-39. doi: 10.1006/plas.2001.1534. Plasmid. 2001. PMID: 11591138
-
KpnAI, a new type I restriction-modification system in Klebsiella pneumoniae.J Mol Biol. 1997 Aug 22;271(3):342-8. doi: 10.1006/jmbi.1997.1202. J Mol Biol. 1997. PMID: 9268663
-
Positive selection, cloning vectors for gram-positive bacteria based on a restriction endonuclease cassette.Plasmid. 1996 Jan;35(1):37-45. doi: 10.1006/plas.1996.0004. Plasmid. 1996. PMID: 8693025
-
EcoR124I: from plasmid-encoded restriction-modification system to nanodevice.Microbiol Mol Biol Rev. 2008 Jun;72(2):365-77, table of contents. doi: 10.1128/MMBR.00043-07. Microbiol Mol Biol Rev. 2008. PMID: 18535150 Free PMC article. Review.
-
[Genetic determinants of restriction-modification systems in bacteria].Mol Gen Mikrobiol Virusol. 1990 Jul;(7):3-11. Mol Gen Mikrobiol Virusol. 1990. PMID: 2215521 Review. Russian.
Cited by
-
Transcriptome analyses of cells carrying the Type II Csp231I restriction-modification system reveal cross-talk between two unrelated transcription factors: C protein and the Rac prophage repressor.Nucleic Acids Res. 2019 Oct 10;47(18):9542-9556. doi: 10.1093/nar/gkz665. Nucleic Acids Res. 2019. PMID: 31372643 Free PMC article.
-
Natural C-independent expression of restriction endonuclease in a C protein-associated restriction-modification system.Nucleic Acids Res. 2016 Apr 7;44(6):2646-60. doi: 10.1093/nar/gkv1331. Epub 2015 Dec 9. Nucleic Acids Res. 2016. PMID: 26656489 Free PMC article.
-
Molecular basis for lethal cross-talk between two unrelated bacterial transcription factors - the regulatory protein of a restriction-modification system and the repressor of a defective prophage.Nucleic Acids Res. 2022 Oct 28;50(19):10964-10980. doi: 10.1093/nar/gkac914. Nucleic Acids Res. 2022. PMID: 36271797 Free PMC article.
-
Natural tuning of restriction endonuclease synthesis by cluster of rare arginine codons.Sci Rep. 2019 Apr 9;9(1):5808. doi: 10.1038/s41598-019-42311-w. Sci Rep. 2019. PMID: 30967604 Free PMC article.
-
Molecular Characterization of a Restriction Endonuclease PsaI from Pseudomonas anguilliseptica KM9 and Sequence Analysis of the PsaI R-M System.Int J Mol Sci. 2025 Jul 8;26(14):6548. doi: 10.3390/ijms26146548. Int J Mol Sci. 2025. PMID: 40724798 Free PMC article.
References
-
- Allen, M. J., A. Collick, and A. J. Jeffreys. 1994. Use of vectorette and subvectorette PCR to isolate transgene flanking DNA. PCR Methods Appl. 4:71-75. - PubMed
-
- Anton, B. P., D. F. Heiter, J. S. Benner, E. J. Hess, L. Greenough, L. S. Moran, B. E. Slatko, and J. E. Brooks. 1997. Cloning and characterization of the Bg/II restriction-modification system reveals a possible evolutionary footprint. Gene 187:19-27. - PubMed
-
- Arnold, C., and I. J. Hodgson. 1991. Vectorette PCR: a novel approach to genomic walking. PCR Methods Appl. 1:39-42. - PubMed
-
- Bart, A., J. Dankert, and A. van der Ende. 1999. Operator sequences for the regulatory proteins of restriction modification systems. Mol. Microbiol. 31:1277-1278. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

