Searching genomes for ribozymes and riboswitches
- PMID: 17472738
- PMCID: PMC1895996
- DOI: 10.1186/gb-2007-8-4-210
Searching genomes for ribozymes and riboswitches
Abstract
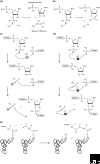
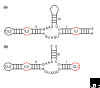
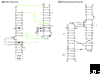
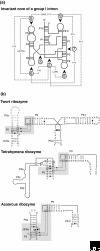
New regulatory RNAs with complex structures have recently been discovered, among them the first catalytic riboswitch, a gene-regulatory RNA sequence with catalytic activity. Here we discuss some of the experimental approaches and theoretical difficulties attached to the identification of new ribozymes in genomes.
Figures

References
-
- Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. doi: 10.1038/323349a0. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

