Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
- PMID: 17473007
- PMCID: PMC2206663
- DOI: 10.1110/ps.072872007
Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Abstract
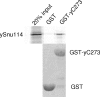
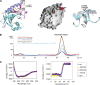
Prp8 is a critical pre-mRNA splicing factor. Prp8 is proposed to help form and stabilize the spliceosome catalytic core and to be an important regulator of spliceosome activation. Mutations in human Prp8 (hPrp8) cause a severe form of the genetic disorder retinitis pigmentosa, RP13. Understanding the molecular mechanism of Prp8's function in pre-mRNA splicing and RP13 has been hindered by its large size (over 2000 amino acids) and remarkably low-sequence similarity with other proteins. Here we present the crystal structure of the C-terminal domain (the last 273 residues) of Caenorhabditis elegans Prp8 (cPrp8). The core of the C-terminal domain is an alpha/beta structure that forms the MPN (Mpr1, Pad1 N-terminal) fold but without Zn(2+) coordination. We propose that the C-terminal domain is a protein interaction domain instead of a Zn(2+)-dependent metalloenzyme as proposed for some MPN proteins. Mapping of RP13 mutants on the Prp8 structure suggests that these residues constitute a binding surface between Prp8 and other partner(s), and the disruption of this interaction provides a plausible molecular mechanism for RP13.
Figures


Similar articles
-
Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa.Mol Cell. 2007 Feb 23;25(4):615-24. doi: 10.1016/j.molcel.2007.01.023. Mol Cell. 2007. PMID: 17317632
-
Prp8 retinitis pigmentosa mutants cause defects in the transition between the catalytic steps of splicing.RNA. 2016 May;22(5):793-809. doi: 10.1261/rna.055459.115. Epub 2016 Mar 11. RNA. 2016. PMID: 26968627 Free PMC article.
-
A Drosophila model to study retinitis pigmentosa pathology associated with mutations in the core splicing factor Prp8.Dis Model Mech. 2020 Jun 26;13(6):dmm043174. doi: 10.1242/dmm.043174. Dis Model Mech. 2020. PMID: 32424050 Free PMC article.
-
Prp8 protein: at the heart of the spliceosome.RNA. 2005 May;11(5):533-57. doi: 10.1261/rna.2220705. RNA. 2005. PMID: 15840809 Free PMC article. Review.
-
Structural dynamics of the N-terminal domain and the Switch loop of Prp8 during spliceosome assembly and activation.Nucleic Acids Res. 2018 May 4;46(8):3833-3840. doi: 10.1093/nar/gky242. Nucleic Acids Res. 2018. PMID: 29635373 Free PMC article. Review.
Cited by
-
Functions and regulation of the Brr2 RNA helicase during splicing.Cell Cycle. 2016 Dec 16;15(24):3362-3377. doi: 10.1080/15384101.2016.1249549. Epub 2016 Oct 28. Cell Cycle. 2016. PMID: 27792457 Free PMC article. Review.
-
Suppressors of the cdc-25.1(gf)-associated intestinal hyperplasia reveal important maternal roles for prp-8 and a subset of splicing factors in C. elegans.RNA. 2008 Dec;14(12):2618-33. doi: 10.1261/rna.1168408. Epub 2008 Oct 22. RNA. 2008. PMID: 18945809 Free PMC article.
-
Detailed close-ups and the big picture of spliceosomes.Curr Opin Struct Biol. 2008 Jun;18(3):315-20. doi: 10.1016/j.sbi.2008.05.005. Epub 2008 Jun 10. Curr Opin Struct Biol. 2008. PMID: 18550358 Free PMC article. Review.
-
Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability.J Mol Biol. 2011 Oct 21;413(2):416-29. doi: 10.1016/j.jmb.2011.08.029. Epub 2011 Aug 24. J Mol Biol. 2011. PMID: 21888914 Free PMC article.
-
Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site.Structure. 2013 Jun 4;21(6):910-19. doi: 10.1016/j.str.2013.04.017. Structure. 2013. PMID: 23727230 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

