Inhibition of U snRNP assembly by a virus-encoded proteinase
- PMID: 17473171
- PMCID: PMC1855234
- DOI: 10.1101/gad.1535607
Inhibition of U snRNP assembly by a virus-encoded proteinase
Abstract
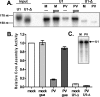
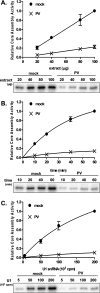
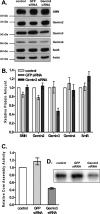
It has been proposed that defects in the assembly of spliceosomal uridine-rich small nuclear ribonucleoprotein (U snRNP) complexes could account for the death of motor neurons in spinal muscular atrophy (SMA). We discovered that infection of cultured cells with poliovirus results in the specific cleavage of the host factor Gemin3 by a virus-encoded proteinase, 2A(pro). Gemin3 is a component of the macromolecular SMN complex that mediates assembly of U snRNP complexes by aiding the heptameric oligomerization of Sm proteins onto U snRNAs. Using in vitro Sm core assembly assays, we found that lowering the intracellular amounts of Gemin3 by either poliovirus infection or small interfering RNA (siRNA)-mediated knockdown of Gemin3 resulted in reduced assembly of U snRNPs. Immunofluorescence analyses revealed a specific redistribution of Sm proteins from the nucleoplasm to the cytoplasmic periphery of the nucleus in poliovirus-infected cells. We propose that defects in U snRNP assembly may be shared features of SMA and poliomyelitis.
Figures
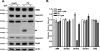


References
-
- Baccon J., Pellizzoni L., Rappsilber J., Mann M., Dreyfuss G., Pellizzoni L., Rappsilber J., Mann M., Dreyfuss G., Rappsilber J., Mann M., Dreyfuss G., Mann M., Dreyfuss G., Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 2002;277:31957–31962. - PubMed
-
- Briese M., Esmaeili B., Sattelle D.B., Esmaeili B., Sattelle D.B., Sattelle D.B. Is spinal muscular atrophy the result of defects in motor neuron processes? Bioessays. 2005;27:946–957. - PubMed
-
- Buhler D., Raker V., Luhrmann R., Fischer U., Raker V., Luhrmann R., Fischer U., Luhrmann R., Fischer U., Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: Implications for spinal muscular atrophy. Hum. Mol. Genet. 1999;8:2351–2357. - PubMed
-
- Carissimi C., Saieva L., Baccon J., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Saieva L., Baccon J., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Baccon J., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Sawyer A., Rappsilber J., Pellizzoni L., Rappsilber J., Pellizzoni L., Pellizzoni L. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J. Biol. Chem. 2006;281:8126–8134. - PubMed
-
- Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Bassell G.J., Burghes A.H., Beattie C.E., Burghes A.H., Beattie C.E., Beattie C.E. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 2006;26:11014–11022. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
