An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication
- PMID: 17475638
- PMCID: PMC1933353
- DOI: 10.1128/JVI.00549-07
An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication
Abstract
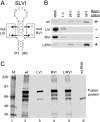
Higher-order cis-acting RNA replication structures have been identified in the 3'- and 5'-terminal untranslated regions (UTRs) of a bovine coronavirus (BCoV) defective interfering (DI) RNA. The UTRs are identical to those in the viral genome, since the 2.2-kb DI RNA is composed of only the two ends of the genome fused between an internal site within the 738-nucleotide (nt) 5'-most coding region (the nsp1, or p28, coding region) and a site just 4 nt upstream of the 3'-most open reading frame (ORF) (the N gene). The joined ends of the viral genome in the DI RNA create a single continuous 1,635-nt ORF, 288 nt of which come from the 738-nt nsp1 coding region. Here, we have analyzed features of the 5'-terminal 288-nt portion of the nsp1 coding region within the continuous ORF that are required for DI RNA replication. We observed that (i) the 5'-terminal 186 nt of the nsp1 coding region are necessary and sufficient for DI RNA replication, (ii) two Mfold-predicted stem-loops within the 186-nt sequence, named SLV (nt 239 to 310) and SLVI (nt 311 to 340), are supported by RNase structure probing and by nucleotide covariation among closely related group 2 coronaviruses, and (iii) SLVI is a required higher-order structure for DI RNA replication based on mutation analyses. The function of SLV has not been evaluated. We conclude that SLVI within the BCoV nsp1 coding region is a higher-order cis-replication element for DI RNA and postulate that it functions similarly in the viral genome.
Figures


References
-
- Beerens, N., and E. J. Snijder. 2006. RNA signals in the 3′ terminus of the genome of equine arteritis virus are required for viral RNA synthesis. J. Gen. Virol. 87:1977-1983. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

