The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA
- PMID: 17475909
- PMCID: PMC1975835
- DOI: 10.1182/blood-2007-02-075184
The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA
Abstract
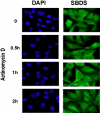
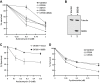
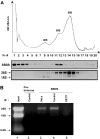
Shwachman-Diamond syndrome (SDS) is an autosomal recessive disorder characterized by bone marrow failure, exocrine pancreatic dysfunction, and leukemia predisposition. Mutations in the SBDS gene are identified in most patients with SDS. SBDS encodes a highly conserved protein of unknown function. Data from SBDS orthologs suggest that SBDS may play a role in ribosome biogenesis or RNA processing. Human SBDS is enriched in the nucleolus, the major cellular site of ribosome biogenesis. Here we report that SBDS nucleolar localization is dependent on active rRNA transcription. Cells from patients with SDS or Diamond-Blackfan anemia are hypersensitive to low doses of actinomycin D, an inhibitor of rRNA transcription. The addition of wild-type SBDS complements the actinomycin D hypersensitivity of SDS patient cells. SBDS migrates together with the 60S large ribosomal subunit in sucrose gradients and coprecipitates with 28S ribosomal RNA (rRNA). Loss of SBDS is not associated with a discrete block in rRNA maturation or with decreased levels of the 60S ribosomal subunit. SBDS forms a protein complex with nucleophosmin, a multifunctional protein implicated in ribosome biogenesis and leukemogenesis. Our studies support the addition of SDS to the growing list of human bone marrow failure syndromes involving the ribosome.
Figures
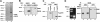
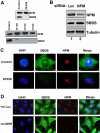
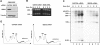

References
-
- Shwachman H, Diamond LK, Oski FA, Khaw KT. The syndrome of pancreatic insufficiency and bone marrow dysfunction. J Pediatr. 1964;65:645–663. - PubMed
-
- Bodian M, Sheldon W, Lightwood R. Congenital hypoplasia of the exocrine pancreas. Acta Paediatr. 1964;53:282–293. - PubMed
-
- Shimamura A. Shwachman-Diamond syndrome. Semin Hematol. 2006;43:178–188. - PubMed
-
- Dror Y. Shwachman-Diamond syndrome. Pediatr Blood Cancer. 2005;45:892–901. - PubMed
-
- Boocock GR, Morrison JA, Popovic M, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

