Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression
- PMID: 17476304
- PMCID: PMC1894761
- DOI: 10.1038/sj.emboj.7601715
Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression
Abstract
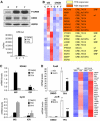
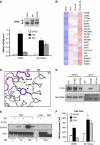
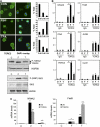
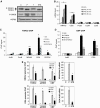
A number of hormones and growth factors stimulate gene expression by promoting the phosphorylation of CREB (P-CREB), thereby enhancing its association with the histone acetylase paralogs p300 and CBP (CBP/p300). Relative to cAMP, stress signals trigger comparable amounts of CREB phosphorylation, but have minimal effects on CRE-dependent transcription. Here, we show that the latent cytoplasmic coactivator TORC2 mediates target gene activation in response to cAMP signaling by associating with CBP/p300 and increasing its recruitment to a subset of CREB target genes. TORC2 is not activated in response to stress signals, however; and in its absence, P-CREB is unable to stimulate CRE-dependent transcription, due to a block in CBP recruitment. The effect of TORC2 on CBP/p300 promoter occupancy appears pivotal because a gain of function mutant CREB polypeptide with increased affinity for CBP restored CRE-mediated transcription in cells exposed to stress signals. Taken together, these results indicate that TORC2 is one of the long sought after cofactors that mediates the differential effects of cAMP and stress pathways on CREB target gene expression.
Figures

Similar articles
-
Temporal recruitment of transcription factors at the 3',5'-cyclic adenosine 5'-monophosphate-response element of the human GnRH-II promoter.Endocrinology. 2008 Oct;149(10):5162-71. doi: 10.1210/en.2008-0481. Epub 2008 Jul 3. Endocrinology. 2008. PMID: 18599546
-
Molecular Basis for the Mechanism of Constitutive CBP/p300 Coactivator Recruitment by CRTC1-MAML2 and Its Implications in cAMP Signaling.Biochemistry. 2015 Sep 8;54(35):5439-46. doi: 10.1021/acs.biochem.5b00332. Epub 2015 Aug 21. Biochemistry. 2015. PMID: 26274502 Free PMC article.
-
Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB.Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10936-41. doi: 10.1073/pnas.191152098. Epub 2001 Sep 4. Proc Natl Acad Sci U S A. 2001. PMID: 11535812 Free PMC article.
-
Latent viruses can cause disease by disrupting the competition for the limiting factor p300/CBP.Cell Mol Biol Lett. 2018 Nov 26;23:56. doi: 10.1186/s11658-018-0121-1. eCollection 2018. Cell Mol Biol Lett. 2018. PMID: 30505323 Free PMC article. Review.
-
p300/CBP as a Key Nutritional Sensor for Hepatic Energy Homeostasis and Liver Fibrosis.Biomed Res Int. 2018 May 15;2018:8168791. doi: 10.1155/2018/8168791. eCollection 2018. Biomed Res Int. 2018. PMID: 29862292 Free PMC article. Review.
Cited by
-
Glucocorticoids Alter CRTC-CREB Signaling in Muscle Cells: Impact on PGC-1α Expression and Atrophy Markers.PLoS One. 2016 Jul 12;11(7):e0159181. doi: 10.1371/journal.pone.0159181. eCollection 2016. PLoS One. 2016. PMID: 27404111 Free PMC article.
-
Anabolic and Pro-metabolic Functions of CREB-CRTC in Skeletal Muscle: Advantages and Obstacles for Type 2 Diabetes and Cancer Cachexia.Front Endocrinol (Lausanne). 2019 Aug 2;10:535. doi: 10.3389/fendo.2019.00535. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 31428057 Free PMC article. Review.
-
A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway.Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20314-9. doi: 10.1073/pnas.0707999105. Epub 2007 Dec 11. Proc Natl Acad Sci U S A. 2007. PMID: 18077367 Free PMC article.
-
Hormonal regulation of gluconeogenic gene transcription in the liver.J Biosci. 2010 Sep;35(3):473-84. doi: 10.1007/s12038-010-0052-0. J Biosci. 2010. PMID: 20826956 Review.
-
Double null cells reveal that CBP and p300 are dispensable for p53 targets p21 and Mdm2 but variably required for target genes of other signaling pathways.Cell Cycle. 2011 Jan 15;10(2):212-21. doi: 10.4161/cc.10.2.14542. Epub 2011 Jan 15. Cell Cycle. 2011. PMID: 21220944 Free PMC article.
References
-
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M (2004) Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol 14: 2156–2161 - PubMed
-
- Bonni A, Ginty D, Dudek H, Greenberg M (1995) Serine 133 phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol Cell Neurosci 6: 168–183 - PubMed
-
- Brindle P, Linke S, Montminy M (1993) Analysis of a PK-A dependent activator in CREB reveals a new role for the CREM Family of repressors. Nature 364: 821–824 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

