Building epidemiological models from R0: an implicit treatment of transmission in networks
- PMID: 17476770
- PMCID: PMC1766386
- DOI: 10.1098/rspb.2006.0057
Building epidemiological models from R0: an implicit treatment of transmission in networks
Abstract
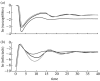
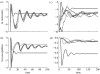
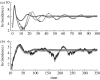
Simple deterministic models are still at the core of theoretical epidemiology despite the increasing evidence for the importance of contact networks underlying transmission at the individual level. These mean-field or 'compartmental' models based on homogeneous mixing have made, and continue to make, important contributions to the epidemiology and the ecology of infectious diseases but fail to reproduce many of the features observed for disease spread in contact networks. In this work, we show that it is possible to incorporate the important effects of network structure on disease spread with a mean-field model derived from individual level considerations. We propose that the fundamental number known as the basic reproductive number of the disease, R0, which is typically derived as a threshold quantity, be used instead as a central parameter to construct the model from. We show that reliable estimates of individual level parameters can replace a detailed knowledge of network structure, which in general may be difficult to obtain. We illustrate the proposed model with small world networks and the classical example of susceptible-infected-recovered (SIR) epidemics.
Figures

Similar articles
-
Epidemic thresholds in dynamic contact networks.J R Soc Interface. 2009 Mar 6;6(32):233-41. doi: 10.1098/rsif.2008.0218. J R Soc Interface. 2009. PMID: 18664429 Free PMC article.
-
Susceptible-infectious-recovered models revisited: from the individual level to the population level.Math Biosci. 2014 Apr;250:26-40. doi: 10.1016/j.mbs.2014.02.001. Epub 2014 Feb 12. Math Biosci. 2014. PMID: 24530806
-
Susceptible-infected-recovered epidemics in dynamic contact networks.Proc Biol Sci. 2007 Dec 7;274(1628):2925-33. doi: 10.1098/rspb.2007.1159. Proc Biol Sci. 2007. PMID: 17878137 Free PMC article.
-
Perspectives on the basic reproductive ratio.J R Soc Interface. 2005 Sep 22;2(4):281-93. doi: 10.1098/rsif.2005.0042. J R Soc Interface. 2005. PMID: 16849186 Free PMC article. Review.
-
Networks and epidemic models.J R Soc Interface. 2005 Sep 22;2(4):295-307. doi: 10.1098/rsif.2005.0051. J R Soc Interface. 2005. PMID: 16849187 Free PMC article. Review.
Cited by
-
Epidemic protection zones: centred on cases or based on connectivity?Transbound Emerg Dis. 2012 Oct;59(5):464-9. doi: 10.1111/j.1865-1682.2011.01301.x. Epub 2012 Feb 24. Transbound Emerg Dis. 2012. PMID: 22360843 Free PMC article.
-
When individual behaviour matters: homogeneous and network models in epidemiology.J R Soc Interface. 2007 Oct 22;4(16):879-91. doi: 10.1098/rsif.2007.1100. J R Soc Interface. 2007. PMID: 17640863 Free PMC article.
-
A paradox of epidemics between the state and parameter spaces.Sci Rep. 2018 May 14;8(1):7517. doi: 10.1038/s41598-018-25931-6. Sci Rep. 2018. PMID: 29760412 Free PMC article.
-
A unified framework of immunological and epidemiological dynamics for the spread of viral infections in a simple network-based population.Theor Biol Med Model. 2007 Dec 20;4:49. doi: 10.1186/1742-4682-4-49. Theor Biol Med Model. 2007. PMID: 18096067 Free PMC article.
-
Simple models for complex systems: exploiting the relationship between local and global densities.Theor Ecol. 2011;4(2):211-222. doi: 10.1007/s12080-011-0116-2. Epub 2011 Mar 11. Theor Ecol. 2011. PMID: 25540675 Free PMC article.
References
-
- Anderson R.M, May R.M. Spatial, temporal, and genetic heterogeneity in host populations and the design of immunization programmes. Institute of Mathematics and its Applications. J. Math. Appl. Med. Biol. 1984;1:233–266. - PubMed
-
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1992. Infectious diseases of humans.
-
- Andersson H. Epidemics in a population with social structure. Math. Biosci. 1997;140:79–84. doi:10.1016/S0025-5564(96)00129-0 - DOI - PubMed
-
- Aparicio J.P, Capurro A.F, Castillo-Chavez C. Transmission and dynamics of tuberculosis on generalized households. J. Theor. Biol. 2000;206:327–341. doi:10.1006/jtbi.2000.2129 - DOI - PubMed
-
- Barabási A.-L. Perseus; Cambridge, MA: 2002. Linked: the new science of networks.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

