Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143
- PMID: 17478512
- PMCID: PMC1904299
- DOI: 10.1093/nar/gkm239
Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143
Abstract
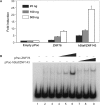
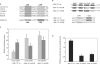
BubR1 is a key protein mediating spindle checkpoint activation. Loss of this checkpoint control results in chromosomal instability and aneuploidy. The transcriptional regulation of the cell cycle regulated human BUB1B gene, which encodes BubR1, was investigated in this report. A minimal BUB1B gene promoter containing 464 bp upstream from the translation initiation codon was sufficient for cell cycle regulated promoter activity. A pivotal role for transcription factor hStaf/ZNF143 in the expression of the BUB1B gene was demonstrated through gel retardation assays, transient expression of mutant BUB1B promoter-reporter gene constructs and chromatin immunoprecipitation assay. Two phylogenetically conserved hStaf/ZNF143-binding sites (SBS) were identified which are indispensable for BUB1B promoter activity. In addition, we found that the domain covering the transcription start sites contains conserved boxes homologous to initiator (Inr), cell cycle dependent (CDE) and cell cycle genes homology regions (CHR) elements. Mutations within the CDE and CHR elements led to diminished cell cycle regulation of BUB1B transcription. These results demonstrate that BUB1B gene transcription is positively regulated by hStaf/ZNF143, a ubiquitously expressed factor, and that the CDE-CHR tandem element was essential for G2/M-specific transcription of the BUB1B gene.
Figures




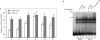
Similar articles
-
Transcription factor hStaf/ZNF143 is required for expression of the human TFAM gene.Gene. 2007 Oct 15;401(1-2):145-53. doi: 10.1016/j.gene.2007.07.011. Epub 2007 Jul 21. Gene. 2007. PMID: 17707600
-
The cancer antiapoptosis mouse survivin gene: characterization of locus and transcriptional requirements of basal and cell cycle-dependent expression.Cancer Res. 1999 Jul 1;59(13):3143-51. Cancer Res. 1999. PMID: 10397257
-
The proximal promoter of the human cathepsin G gene conferring myeloid-specific expression includes C/EBP, c-myb and PU.1 binding sites.Gene. 2005 Aug 15;356:193-202. doi: 10.1016/j.gene.2005.05.004. Gene. 2005. PMID: 16019164
-
The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription.FEBS J. 2010 Feb;277(4):877-93. doi: 10.1111/j.1742-4658.2009.07508.x. Epub 2009 Dec 15. FEBS J. 2010. PMID: 20015071 Review.
-
[Determination of transcription start sites: CAGE & GIS tag sequences].Tanpakushitsu Kakusan Koso. 2004 Dec;49(17 Suppl):2701-3. Tanpakushitsu Kakusan Koso. 2004. PMID: 15669242 Review. Japanese. No abstract available.
Cited by
-
Prognostic Signature for Lung Adenocarcinoma Patients Based on Cell-Cycle-Related Genes.Front Cell Dev Biol. 2021 Mar 18;9:655950. doi: 10.3389/fcell.2021.655950. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869220 Free PMC article.
-
Experimental and clinical evidence suggests that GRPEL2 plays an oncogenic role in HCC development.Am J Cancer Res. 2021 Sep 15;11(9):4175-4198. eCollection 2021. Am J Cancer Res. 2021. PMID: 34659882 Free PMC article.
-
Widespread Dysregulation of Long Noncoding Genes Associated With Fatty Acid Metabolism, Cell Division, and Immune Response Gene Networks in Xenobiotic-exposed Rat Liver.Toxicol Sci. 2020 Apr 1;174(2):291-310. doi: 10.1093/toxsci/kfaa001. Toxicol Sci. 2020. PMID: 31926019 Free PMC article.
-
Pan-cancer investigation of psoriasis-related BUB1B gene: genetical alteration and oncogenic immunology.Sci Rep. 2023 Apr 13;13(1):6058. doi: 10.1038/s41598-023-33174-3. Sci Rep. 2023. PMID: 37055476 Free PMC article.
-
Integrated Network Analysis Decipher ZNF384-Related miR-20b-5p and miR-424-5p in Colon Adenocarcinoma.Cancer Rep (Hoboken). 2025 May;8(5):e70233. doi: 10.1002/cnr2.70233. Cancer Rep (Hoboken). 2025. PMID: 40405535 Free PMC article.
References
-
- Lengauer C. Cancer. An unstable liaison. Science. 2003;300:442–443. - PubMed
-
- Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. - PubMed
-
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. - PubMed
-
- Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, Mahmood R, Yang YM, Xu M, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

