Transcription-coupled deposition of histone modifications during MHC class II gene activation
- PMID: 17478518
- PMCID: PMC1904273
- DOI: 10.1093/nar/gkm214
Transcription-coupled deposition of histone modifications during MHC class II gene activation
Abstract
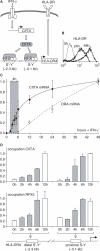
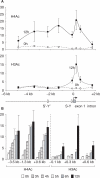
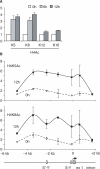
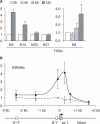
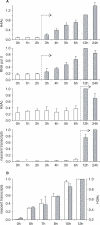
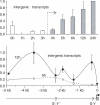
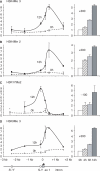
Posttranslational histone modifications associated with actively expressed genes are generally believed to be introduced primarily by histone-modifying enzymes that are recruited by transcription factors or their associated co-activators. We have performed a comprehensive spatial and temporal analyses of the histone modifications that are deposited upon activation of the MHC class II gene HLA-DRA by the co-activator CIITA. We find that transcription-associated histone modifications are introduced during two sequential phases. The first phase precedes transcription initiation and is characterized exclusively by a rapid increase in histone H4 acetylation over a large upstream domain. All other modifications examined, including the acetylation and methylation of several residues in histone H3, are restricted to short regions situated at or within the 5' end of the gene and are established during a second phase that is concomitant with ongoing transcription. This second phase is completely abrogated when elongation by RNA polymerase II is blocked. These results provide strong evidence that transcription elongation can play a decisive role in the deposition of histone modification patterns associated with inducible gene activation.
Figures

Similar articles
-
CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter.Nat Immunol. 2001 Jul;2(7):652-7. doi: 10.1038/89810. Nat Immunol. 2001. PMID: 11429551
-
X box-like sequences in the MHC class II region maintain regulatory function.J Immunol. 2005 Jul 15;175(2):1030-40. doi: 10.4049/jimmunol.175.2.1030. J Immunol. 2005. PMID: 16002703
-
Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by Trichostatin A.Nucleic Acids Res. 2006 Feb 1;34(3):765-72. doi: 10.1093/nar/gkj462. Print 2006. Nucleic Acids Res. 2006. PMID: 16452299 Free PMC article.
-
Histone acetylation and methylation: combinatorial players for transcriptional regulation.Subcell Biochem. 2007;41:351-69. Subcell Biochem. 2007. PMID: 17484136 Review.
-
Epigenetic interplay between histone modifications and DNA methylation in gene silencing.Mutat Res. 2008 Jul-Aug;659(1-2):40-8. doi: 10.1016/j.mrrev.2008.02.004. Epub 2008 Feb 29. Mutat Res. 2008. PMID: 18407786 Review.
Cited by
-
CTCF binds to sites in the major histocompatibility complex that are rapidly reconfigured in response to interferon-gamma.Nucleic Acids Res. 2012 Jul;40(12):5262-70. doi: 10.1093/nar/gks158. Epub 2012 Feb 25. Nucleic Acids Res. 2012. PMID: 22367884 Free PMC article.
-
Multiple histone methyl and acetyltransferase complex components bind the HLA-DRA gene.PLoS One. 2012;7(5):e37554. doi: 10.1371/journal.pone.0037554. Epub 2012 May 31. PLoS One. 2012. PMID: 22701520 Free PMC article.
-
Identification of CIITA regulated genetic module dedicated for antigen presentation.PLoS Genet. 2008 Apr 25;4(4):e1000058. doi: 10.1371/journal.pgen.1000058. PLoS Genet. 2008. PMID: 18437201 Free PMC article.
-
The MHC Class II Transactivator CIITA: Not (Quite) the Odd-One-Out Anymore among NLR Proteins.Int J Mol Sci. 2021 Jan 22;22(3):1074. doi: 10.3390/ijms22031074. Int J Mol Sci. 2021. PMID: 33499042 Free PMC article. Review.
-
Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes.Epigenetics. 2014 Feb;9(2):276-85. doi: 10.4161/epi.27021. Epub 2013 Nov 8. Epigenetics. 2014. PMID: 24213554 Free PMC article.
References
-
- Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 2003;4:276–284. - PubMed
-
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. - PubMed
-
- Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci. 2006;11:344–355. - PubMed
-
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. - PubMed
-
- Wright KL, Ting JP. Epigenetic regulation of MHC-II and CIITA genes. Trends Immunol. 2006;27:405–412. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

