Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization
- PMID: 17481394
- PMCID: PMC1963441
- DOI: 10.1016/j.neuron.2007.03.029
Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization
Abstract
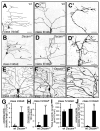
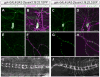
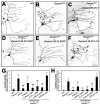
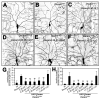
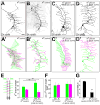
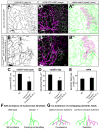
A neuron's dendrites typically do not cross one another. This intrinsic self-avoidance mechanism ensures unambiguous processing of sensory or synaptic inputs. Moreover, some neurons respect the territory of others of the same type, a phenomenon known as tiling. Different types of neurons, however, often have overlapping dendritic fields. We found that Down's syndrome Cell Adhesion Molecule (Dscam) is required for dendritic self-avoidance of all four classes of Drosophila dendritic arborization (da) neurons. However, neighboring mutant class IV da neurons still exhibited tiling, suggesting that self-avoidance and tiling differ in their recognition and repulsion mechanisms. Introducing 1 of the 38,016 Dscam isoforms to da neurons in Dscam mutants was sufficient to significantly restore self-avoidance. Remarkably, expression of a common Dscam isoform in da neurons of different classes prevented their dendrites from sharing the same territory, suggesting that coexistence of dendritic fields of different neuronal classes requires divergent expression of Dscam isoforms.
Figures



Comment in
-
Avoiding the SCAMs.Neuron. 2007 May 3;54(3):350-2. doi: 10.1016/j.neuron.2007.04.018. Neuron. 2007. PMID: 17481387
References
-
- Chen BE, Kondo M, Garnier A, Watson FL, Puettmann-Holgado R, Lamar DR, Schmucker D. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. - PubMed
-
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

