Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis
- PMID: 17482871
- PMCID: PMC7172271
- DOI: 10.1016/j.jcv.2007.03.012
Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis
Abstract
Background: Human rhinoviruses (HRVs) are some of the earliest identified and most commonly detected viruses associated with acute respiratory tract infections (ARTIs) and yet the molecular epidemiology and genomic variation of individual serotypes remains undefined.
Objectives: To molecularly characterise a novel HRV and determine its prevalence and clinical impact on a predominantly paediatric population.
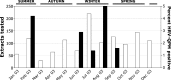
Study design: Nucleotide sequencing was employed to determine the complete HRV-QPM coding sequence. Two novel real-time RT-PCR diagnostic assays were designed and employed to retrospectively screen a well-defined population of 1244 specimen extracts to identify the prevalence of HRV-QPM during 2003.
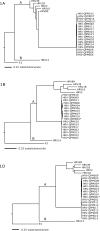
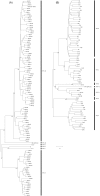
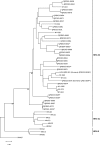
Results: Phylogenetic studies of complete coding sequences defined HRV-QPM as a novel member the genus Rhinovirus residing within the previously described HRV-A2 sub-lineage. Investigation of the relatively short VP1 sequence suggest that the virus is resistant to Pleconaril, setting it apart from the HRV A species. Sixteen additional HRV-QPM strains were detected (1.4% of specimens) often as the sole micro-organism present among infants with suspected bronchiolitis. HRV-QPM was also detected in Europe during 2006, and a closely related virus circulated in the United States during 2004.
Conclusions: We present the molecular characterisation and preliminary clinical impact of a newly identified HRV along with sequences representing additional new HRVs.
Figures

References
-
- Caro V., Guillot S., Delpeyroux F., Crainic R. Molecular strategy for ‘serotyping’ of human enteroviruses. J Gen Virol. 2001;82:79–91. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

