Detecting ultraviolet damage in single DNA molecules by atomic force microscopy
- PMID: 17483180
- PMCID: PMC1948057
- DOI: 10.1529/biophysj.107.108209
Detecting ultraviolet damage in single DNA molecules by atomic force microscopy
Abstract
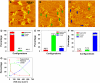
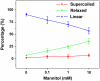
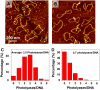
We report detection and quantification of ultraviolet (UV) damage in DNA at a single molecule level by atomic force microscopy (AFM). By combining the supercoiled plasmid relaxation assay with AFM imaging, we find that high doses of medium wave ultraviolet (UVB) and short wave ultraviolet (UVC) light not only produce cyclobutane pyrimidine dimers (CPDs) as reported but also cause significant DNA degradation. Specifically, 12.5 kJ/m(2) of UVC and 165 kJ/m(2) of UVB directly relax 95% and 78% of pUC18 supercoiled plasmids, respectively. We also use a novel combination of the supercoiled plasmid assay with T4 Endonuclease V treatment of irradiated plasmids and AFM imaging of their relaxation to detect damage caused by low UVB doses, which on average produced approximately 0.5 CPD per single plasmid. We find that at very low UVB doses, the relationship between the number of CPDs and UVB dose is almost linear, with 4.4 CPDs produced per Mbp per J/m(2) of UVB radiation. We verified these AFM results by agarose gel electrophoresis separation of UV-irradiated and T4 Endonuclease V treated plasmids. Our AFM and gel electrophoresis results are consistent with the previous result obtained using other traditional DNA damage detection methods. We also show that damage detection assay sensitivity increases with plasmid size. In addition, we used photolyase to mark the sites of UV lesions in supercoiled plasmids for detection and quantification by AFM, and these results were found to be consistent with the results obtained by the plasmid relaxation assay. Our results suggest that AFM can supplement traditional methods for high resolution measurements of UV damage to DNA.
Figures




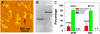
References
-
- Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA Repair and Mutagenesis. ASM Press, Washington, DC.
-
- Sinha, R. P., and D. P. Hader. 2002. UV-induced DNA damage and repair: a review. Photochem. Photobiol. Sci. 1:225–236. - PubMed
-
- Setlow, R. B., and W. L. Carrier. 1966. Pyrimidine dimers in ultraviolet-irradiated DNA's. J. Mol. Biol. 17:237–254. - PubMed
-
- Douki, T., A. Reynaud-Angelin, J. Cadet, and E. Sage. 2003. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 42:9221–9226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

