Mutational analysis of the ompA promoter from Flavobacterium johnsoniae
- PMID: 17483221
- PMCID: PMC1951883
- DOI: 10.1128/JB.00401-07
Mutational analysis of the ompA promoter from Flavobacterium johnsoniae
Abstract
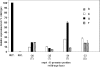
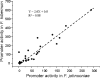
Sequences that mediate the initiation of transcription in Flavobacterium species are not well known. The majority of identified Flavobacterium promoter elements show homology to those of other members of the phylum Bacteroidetes, but not of proteobacteria, and they function poorly in Escherichia coli. In order to analyze the Flavobacterium promoter structure systematically, we investigated the -33 consensus element, -7 consensus element, and spacer length of the Flavobacterium ompA promoter by measuring the effects of site-directed mutations on promoter activity. The nonconserved sequences in the spacer region and in regions close to the consensus motifs were randomized in order to determine their importance for promoter activity. Most of the base substitutions in these regions caused large decreases in promoter activity. The optimal -33/-7 motifs (TTTG/TANNTTTG) were identical to Bacteroides fragilis sigma(ABfr) consensus -33/-7 promoter elements but lacked similarity to the E. coli sigma(70) promoter elements. The length of the spacer separating the -33 and -7 motifs of the ompA promoter also had a pronounced effect on promoter activity, with 19 bp being optimal. In addition to the consensus promoter elements and spacer length, the GC content of the core promoter sequences had a pronounced effect on Flavobacterium promoter activity. This information was used to conduct a scan of the Flavobacterium johnsoniae and B. fragilis genomes for putative promoters, resulting in 188 hits in B. fragilis and 109 hits in F. johnsoniae.
Figures


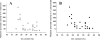
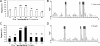
References
-
- Bannantine, J. P., R. G. Barletta, C. O. Thoen, and R. E. Andrews, Jr. 1997. Identification of Mycobacterium paratuberculosis gene expression signals. Microbiology 143:921-928. - PubMed
-
- Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

